Difference between revisions of "Projects:DiffusionGraphBasedConnectivity"
(Created page with ' Back to UNC Algorithms __NOTOC__ = Muti-directional graph propagation for Diffusion Imaging based Connectivity = This project focuses on connectivity measur…') |
|||
| Line 9: | Line 9: | ||
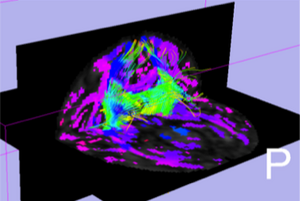
[[Image:UNC_GraphbasedConnectivity_Ex1.png|thumb|300px|Slicer visualization of genu connectivity map overlaid on streamline genu tracts]] | [[Image:UNC_GraphbasedConnectivity_Ex1.png|thumb|300px|Slicer visualization of genu connectivity map overlaid on streamline genu tracts]] | ||
| + | Regional connectivity measurements derived from diffusion imaging datasets are of considerable interest in the neuroimaging community for better understanding white matter connectivity. Current connectivity measurements are usually either based on fiber tractography applied in a Monte-Carlo fashion or are variations of the Hamilton-Jacobi approach. | ||
| + | |||
| + | We propose a novel, graph-based algorithm that provides a fully deterministic, efficient and stable connectivity measure. This method handles crossing fibers and deals well with multiple seed regions. The computation is based on a multi-directional graph propagation algorithm applied to sampled orientation distribution functions computed directly from the original diffusion imaging data. A maximum probability and a minimum cost approach are both possible. | ||
| + | |||
| + | So far we have results on synthetic and limited real datasets to illustrate the potential of our method towards subject-specific connectivity measurements performed in an efficient, stable and reproducible manner. Such individual connectivity measurements would be well suited for studies of neuropathology. | ||
= Publications = | = Publications = | ||
Revision as of 22:03, 1 April 2011
Home < Projects:DiffusionGraphBasedConnectivityBack to UNC Algorithms
Muti-directional graph propagation for Diffusion Imaging based Connectivity
This project focuses on connectivity measurements derived from diffusion imaging datasets in order to better understand cortical and subcortical white matter connectivity. Our research employs a novel, multi-directional graph propagation method that performs a fully deterministic, efficient and stable connectivity computation. The method handles crossing fibers and deals well with multiple seed regions. In addition to the analysis of these connectivity measures in describing brain pathology, they can also be used as scalar maps for use in DTI registration.
Description
Regional connectivity measurements derived from diffusion imaging datasets are of considerable interest in the neuroimaging community for better understanding white matter connectivity. Current connectivity measurements are usually either based on fiber tractography applied in a Monte-Carlo fashion or are variations of the Hamilton-Jacobi approach.
We propose a novel, graph-based algorithm that provides a fully deterministic, efficient and stable connectivity measure. This method handles crossing fibers and deals well with multiple seed regions. The computation is based on a multi-directional graph propagation algorithm applied to sampled orientation distribution functions computed directly from the original diffusion imaging data. A maximum probability and a minimum cost approach are both possible.
So far we have results on synthetic and limited real datasets to illustrate the potential of our method towards subject-specific connectivity measurements performed in an efficient, stable and reproducible manner. Such individual connectivity measurements would be well suited for studies of neuropathology.
Publications
Key Investigators
- UNC Algorithms: Ipek Oguz, Alexis Boucharin, Martin Styner