Difference between revisions of "Projects:GroupwiseRegistration"
| Line 1: | Line 1: | ||
| − | Back to [[NA-MIC_Collaborations|NA- | + | Back to [[NA-MIC_Collaborations|NA-MIC_Collaborations]], [[Algorithm:GATech|Georgia Tech Algorithms]] |
| − | = | + | = Optimal Mass Transport Registration = |
| − | + | The aim of this project to provide a computationaly efficient non-rigid/elastic image registration algorithm based on the Optimal Mass Transport theory. We use the Monge-Kantorovich formulation of the Optimal Mass Transport problem and implement the solution proposed by Haker et al. using multi-resolution and multigrid techniques to speed up the convergence. We also leverage the computation power of general purpose graphics processing units available on standard desktop computing machines to exploit the inherent parallelism in our algorithm. | |
| − | = Description = | + | = Description = |
| − | + | The Optimal Mass Transport problem was first formulated by a Frech engineer Gasper Monge in 1781, and was given a modern formulation in the work of Kantorovich and, therefore, is now known as the Monge-Kantorovich problem. We extend the work by Haker et al. who compute the optimal warp from a first order partial differential equation, an improvement over earlier proposed higher order methods and those based on linear programming. We implement the algorithm using a coarse-to-fine strategy resulting in phenomenol improvement in convergence. The algorithm also involves inverting the Laplacian in each iteration, which we perform using multigrid methods for even faster per iteration computation times. This method has a number of distinguishing characteristics: | |
| + | |||
| + | # It is a parameter free method. | ||
| + | # It utillizes all of the grayscale data in both images. | ||
| + | # It is symmetrical; the optimal mapping from image A to image B is the inverse of the optimal mapping from B to A. | ||
| + | # No landmarks need to be specified. | ||
| + | # The minimizer of the functional involved is unique; there are no local minimizers. | ||
| + | # The algorithm is designed to take into account changes in densities that result from changes in area or volume. | ||
''Algorithm'' | ''Algorithm'' | ||
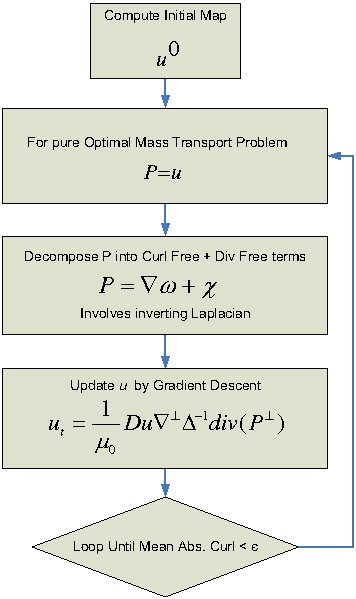
| − | The | + | The flowchart of the algorithm is shown in the following figure. |
| + | |||
| + | [[Image:OMT_Algorithm.jpg| Optimal Mass Transport Algorithm | center]] | ||
| + | |||
| + | <br/> | ||
| − | + | ''Multi-resolution Approach'' | |
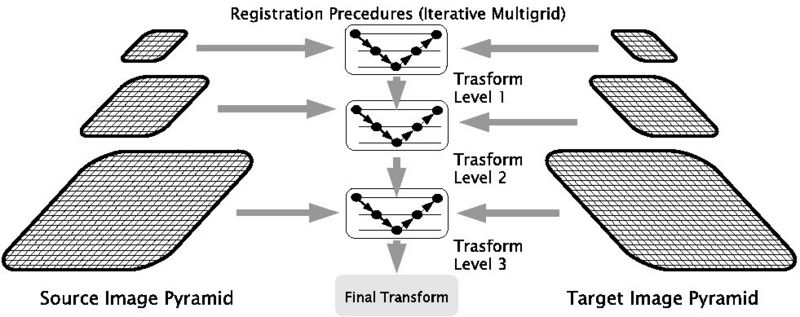
| − | + | Performing image registration using a multi-resolution approach is widely used to improve speed, accuracy and robustness. Registration is first performed at a coarse scale. The spatial mapping determined at coarse scale is then used to initialize registration at the next finer scale. This process is repeated until it reaches the finest scale. Our coarse-to-fine hierarchy comprises of three levels and we use bi-cubic interpolation to interpolate results from coarse to fine grid. This process is depicted in the following figure. | |
| + | <br/> | ||
| − | + | [[Image:Multilevel_diagram.jpg| Multi-resolution Implementation | 800px | center]] | |
''Progress'' | ''Progress'' | ||
| − | + | Below we show the results from registration of two 3D brain MRI datasets. The first data set was pre-operative while the second data set was acquired during surgery (craniotomy). Both were resampled to 256*256*256 for uniform voxel size and the skull was removed. We show the results from two axial slices of the 3D brain volumes. The sag and compression areas can easily be seen in the deformed grid shown below. The reults shown are after 3600 iterations, requiring less than 15 minutes of computation time (Dual Xeon 1.6GHz + nVidia GeForce 8800 GX GPU. The optimal computation time was found to occur for a grid size of 128*128*128 where about 1000 iterations execute in less than 15 seconds. This is due to the memory limitations on the graphics card used. | |
| − | + | * [[Image:Results_brain_sag.JPG | Visual Results | 1000px]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | 3D Registration Results on axial slices. We visualize the optimal transport map (right) in the form of a vector field corresponding to the directions of deformation between pre-op (left) | |
| − | + | and post-op (center) brains. Data size 256*256*256. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ''Project Status'' | |
| − | + | * 2D Multi-resolution Registration using Optimal Mass Transport implemented. | |
| − | + | * 3D Multi-resolution Registration using Optimal Mass Transport of Brain sag datasets implemented. | |
| − | + | * Currently working on validating 3D registration results. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | = Key Investigators = | |
| − | + | * Georgia Tech: Tauseef ur Rehman, Gallagher Pryor, John Melonakos, Allen Tannenbaum | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
= Publications = | = Publications = | ||
| − | + | ''In press'' | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * T. Rehman and A. Tannenbaum. Multigrid Optimal Mass Transport for Image Registration and Morphing. SPIE Computation Imaging V, 2007 | |
| + | * T. Rehman, G. Pryor and A. Tannenbaum. GPU Enhanced Multigrid Optimal Mass Transport for Image Registration and Morphing. Publication in submission. | ||
| + | * T. Rehman, G. Pryor, J. Melonakos and A. Tannenbaum. Multiresolution 3D Nonrigid Registration via Optimal Mass Transport. Publication in submission. | ||
Revision as of 18:51, 9 November 2007
Home < Projects:GroupwiseRegistrationBack to NA-MIC_Collaborations, Georgia Tech Algorithms
Optimal Mass Transport Registration
The aim of this project to provide a computationaly efficient non-rigid/elastic image registration algorithm based on the Optimal Mass Transport theory. We use the Monge-Kantorovich formulation of the Optimal Mass Transport problem and implement the solution proposed by Haker et al. using multi-resolution and multigrid techniques to speed up the convergence. We also leverage the computation power of general purpose graphics processing units available on standard desktop computing machines to exploit the inherent parallelism in our algorithm.
Description
The Optimal Mass Transport problem was first formulated by a Frech engineer Gasper Monge in 1781, and was given a modern formulation in the work of Kantorovich and, therefore, is now known as the Monge-Kantorovich problem. We extend the work by Haker et al. who compute the optimal warp from a first order partial differential equation, an improvement over earlier proposed higher order methods and those based on linear programming. We implement the algorithm using a coarse-to-fine strategy resulting in phenomenol improvement in convergence. The algorithm also involves inverting the Laplacian in each iteration, which we perform using multigrid methods for even faster per iteration computation times. This method has a number of distinguishing characteristics:
- It is a parameter free method.
- It utillizes all of the grayscale data in both images.
- It is symmetrical; the optimal mapping from image A to image B is the inverse of the optimal mapping from B to A.
- No landmarks need to be specified.
- The minimizer of the functional involved is unique; there are no local minimizers.
- The algorithm is designed to take into account changes in densities that result from changes in area or volume.
Algorithm
The flowchart of the algorithm is shown in the following figure.
Multi-resolution Approach
Performing image registration using a multi-resolution approach is widely used to improve speed, accuracy and robustness. Registration is first performed at a coarse scale. The spatial mapping determined at coarse scale is then used to initialize registration at the next finer scale. This process is repeated until it reaches the finest scale. Our coarse-to-fine hierarchy comprises of three levels and we use bi-cubic interpolation to interpolate results from coarse to fine grid. This process is depicted in the following figure.
Progress
Below we show the results from registration of two 3D brain MRI datasets. The first data set was pre-operative while the second data set was acquired during surgery (craniotomy). Both were resampled to 256*256*256 for uniform voxel size and the skull was removed. We show the results from two axial slices of the 3D brain volumes. The sag and compression areas can easily be seen in the deformed grid shown below. The reults shown are after 3600 iterations, requiring less than 15 minutes of computation time (Dual Xeon 1.6GHz + nVidia GeForce 8800 GX GPU. The optimal computation time was found to occur for a grid size of 128*128*128 where about 1000 iterations execute in less than 15 seconds. This is due to the memory limitations on the graphics card used.
3D Registration Results on axial slices. We visualize the optimal transport map (right) in the form of a vector field corresponding to the directions of deformation between pre-op (left) and post-op (center) brains. Data size 256*256*256.
Project Status
- 2D Multi-resolution Registration using Optimal Mass Transport implemented.
- 3D Multi-resolution Registration using Optimal Mass Transport of Brain sag datasets implemented.
- Currently working on validating 3D registration results.
Key Investigators
- Georgia Tech: Tauseef ur Rehman, Gallagher Pryor, John Melonakos, Allen Tannenbaum
Publications
In press
- T. Rehman and A. Tannenbaum. Multigrid Optimal Mass Transport for Image Registration and Morphing. SPIE Computation Imaging V, 2007
- T. Rehman, G. Pryor and A. Tannenbaum. GPU Enhanced Multigrid Optimal Mass Transport for Image Registration and Morphing. Publication in submission.
- T. Rehman, G. Pryor, J. Melonakos and A. Tannenbaum. Multiresolution 3D Nonrigid Registration via Optimal Mass Transport. Publication in submission.