Projects:GroupwiseRegistration
Back to NA-MIC_Collaboration, MGH Algorithms, MIT Algorithms
Groupwise Registration
Image registration is an important tool in medical image analysis. Medical experts traditionally rely on manual comparisons of images, but the abundance of information due to technological advances makes this task a challenging one. We present a fully automatic procedure which achieves correspondence among anatomical structures in medical images. Our registration procedure runs in a simultaneous fashion, with every member of the population approaching the central tendency of the population at the same time. Compared to pairwise algorithms we eliminate the need for selecting a particular reference frame a priori. Therefore, our algorithm can generate unbiased atlases.
Description
A problem associated with the use of pairwise image registration is that the atlas created is dependent on the choice of the reference subject. If there is wide variation within the population the reference subject might not be good representative of the population. Instead of choosing a reference subject and performing pairwise registration, we propose a groupwise non-rigid registration algorithm to simultaneously register all subjects in a population to a common reference coordinate system. By registering all the images simultaneously, the resulting deformation fields contain information about the variability across the population. Using the resulting deformation fields we obtain a dense and consistent correspondence across the population which allows for the creation of population-specific atlases.
Algorithm
The method we propose uses the congealing framework with an information theoretic objective function. The congealing technique was introduced to capture the central tendency of a collection of input images for classification purposes [1]. We adapt the congealing framework to a population of grayscale-valued 3D data volumes and provide a computationally efficient implementation via a stochastic gradient-based optimization procedure in a multi-resolution framework.
In the congealing framework we use the sum of voxel-wise entropies as a joint alignment criterion. If the images are aligned properly, intensity values at corresponding coordinate locations from all the inputs form a low entropy distribution; hence, the objective function achieves a minimum. This statement holds even if the intensity values are not identical. Therefore, an entropy-based objective function makes the algorithm considerably robust to noise and bias fields. As B-Spline based non-rigid deformations with dense control points can overfit the data, we add a term to our objective function penalizing non-affine deformations (similar to [2]).
To find the set of non-rigid transformations that aligns all subjects to a common reference coordinate system, B-Spline control points are overlayed uniformly over the image sets. The directions of movement of the B-Spline control points are found by calculating the gradient of the objective function with respect to the control point displacements. The control points are updated using gradient descents until the objective function converges to a local minimum. While minimizing the objective function we constrain the sum of all the deformations to be zero to force the mean scale of the registered images to be the same as the original images. Therefore, the gradient of the objective function is projected onto the constraint surface after each iteration. To reduce the computational time of the algorithm we make use of stochastic subsampling in a multiresolution setting (similar to that of [3]). To estimate the entropy measure from the samples, we use the Parzen windowing method [4].
Prior to non-rigid registration, affine congealing [5] is performed on the data to obtain a good initialization for B-Splines. Afterwards, B-Spline based nonrigid deformations are used to obtain a dense deformation field on the data set. We embed both registrations in a multiresolution setting to avoid the deformation fields to be trapped by local minima. Our implementation of groupwise non-rigid registration is based on open source Insight Toolkit(ITK) and is publicly available [6].
Progress
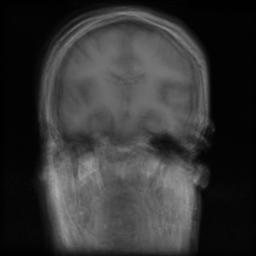
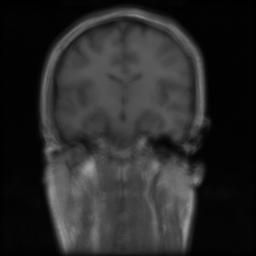
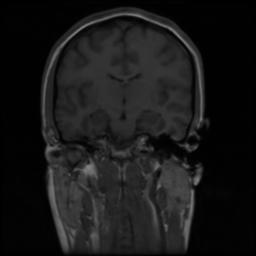
We present the results of our algorithm on two synthetic data sets. The first data set is generated by applying 30 random affine transformations to an MRI data set of size 256x256x128. The original images were recovered by our algorithm. The second data set was created by applying 30 random B-Spline based nonrigid deformations to the same image. As the mean images indicate the original images are obtained accurately.
Software
Our algorithm is implemented using ITK. The source code is publicly available through ITK's svn servers. The project is located under the folder NAMICSandBox/trunk/MultiImageRegistration. [source code]
- Current implementation supports groupwise registration using
- congealing
- registering to the mean
- registering all pairs to all pairs
- joint entropy
- An efficient implementation of B-Splines is provided
- All methods have a fast multithreaded implementation
Publications
- E. Miller, N. Matsakis and P. Viola. Learning from One Example through Shared Densities on Transforms. In IEEE Computer Society Conference on Computer Vision and Pattern Recognition , vol. 1, pp. 464-471, 2000.
- D. Rueckert, L. Sonodo, I.C. Hayes, D.L.G. Hill, M.O. Leach, D.J. Hawkes. Nonrigid Registration Using Free-Form Deformations: Application to Breast MR Images. In IEEE Transactions on Medical Imaging, vol. 18, n. 8, pp. 712-721, 1999.
- W.M. Wells, P. Viola, H. Atsumi, S. Nakajima and R. Kikinis. Multimodality Image Registration by Maximization of Mutual Information. In Medical Image Analysis , vol. 1, n. 1, pp. 35-51, 1996.
- R.O. Duda, P.E. Hart and D.G. Stork. Pattern Classification. New York John Wiley, 2001.
- L. Zollei, E. Learned-Miller, E. Grimson and W. Wells. Efficient Population Registration of 3D Data. In ICCV 2005 , vol. 3765 of LNCS. pp. 291-301, 2005.
- The Insight Segmentation and Registration Toolkit, http://www.itk.org.