Difference between revisions of "Projects:KnowledgeBasedBayesianSegmentation"
| (26 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[NA- | + | Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm:Stony Brook|Stony Brook University Algorithms]], [[Engineering:Kitware|Kitware Engineering]] |
| − | + | __NOTOC__ | |
| − | + | = Knowledge Based Bayesian Segmentation = | |
This ITK filter is a segmentation algorithm which utilizes Bayes's Rule along with an affine-invarient anisotropic smoothing filter. | This ITK filter is a segmentation algorithm which utilizes Bayes's Rule along with an affine-invarient anisotropic smoothing filter. | ||
| − | + | = Description = | |
''Use Case'' | ''Use Case'' | ||
| Line 13: | Line 13: | ||
''Data'' | ''Data'' | ||
| − | We are | + | We have applied this algorithm to 20 normal brain MRI data-sets. We used publicly available data-sets from |
| + | the Internet Brain Segmentation Repository (IBSR) offered by the Massachusetts General Hospital, Center for | ||
| + | Morphometric Analysis. The IBSR data-sets are T1-weighted, 3D coronal brain scans after having been | ||
| + | positionally normalized. Manual expert segmentations for these data-sets are publicly available and represent | ||
| + | the ground truth used in this work. | ||
''Algorithm'' | ''Algorithm'' | ||
| + | |||
| + | This algorithm can be cast in either a static or dynamic framework. In the static framework, the following is the algorithm: | ||
# The user sets the number of distinct classes for segmentation: 'N' | # The user sets the number of distinct classes for segmentation: 'N' | ||
| Line 24: | Line 30: | ||
# Smooth the posterior images for 'm' iterations using an affine-invarient anisotropic smoothing filter and renormalize after each iteration (default, m = 5) <br /> | # Smooth the posterior images for 'm' iterations using an affine-invarient anisotropic smoothing filter and renormalize after each iteration (default, m = 5) <br /> | ||
# Apply maximum a posteriori rule to apply labeling and finalize segmentation | # Apply maximum a posteriori rule to apply labeling and finalize segmentation | ||
| + | |||
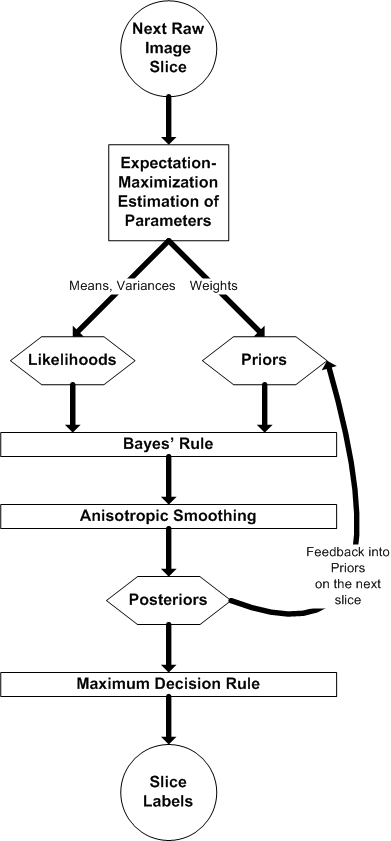
| + | In the dynamic framework, the following image depicts the adaptation of the static framework to the dynamic formulation: | ||
| + | |||
| + | [[Image:Flowchart-classification.png| Dynamic Tissue Tracking Algorithm | center]] | ||
| + | |||
| + | <br/> | ||
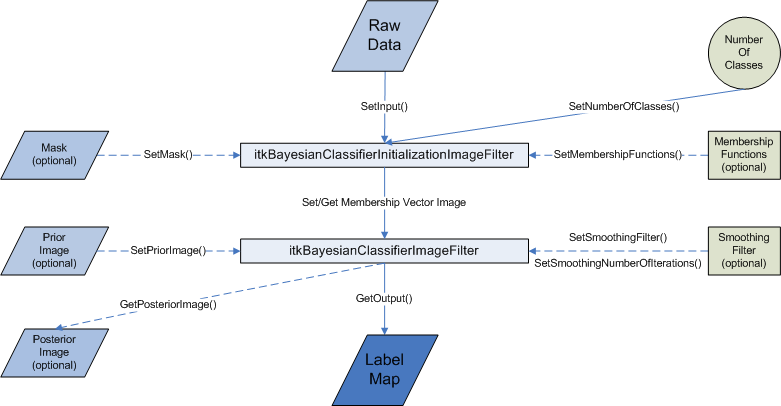
''The ITK filter design'' | ''The ITK filter design'' | ||
| − | + | <br/> | |
| + | |||
| + | [[Image:Flowchart.png| Flowchart]] | ||
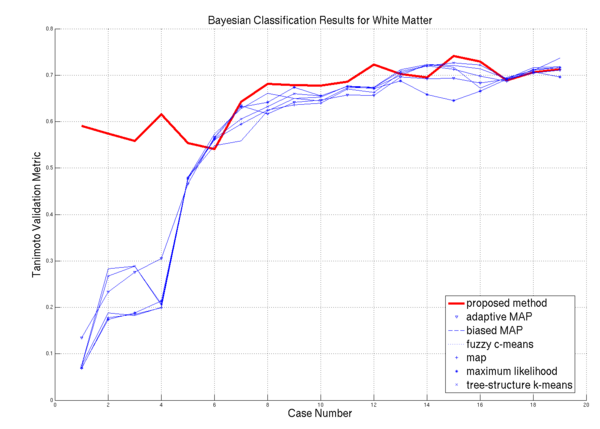
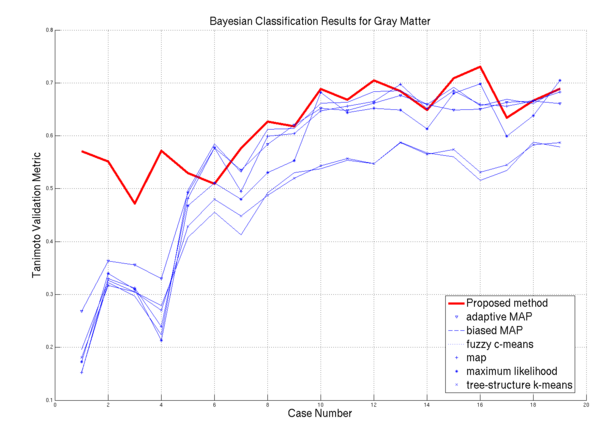
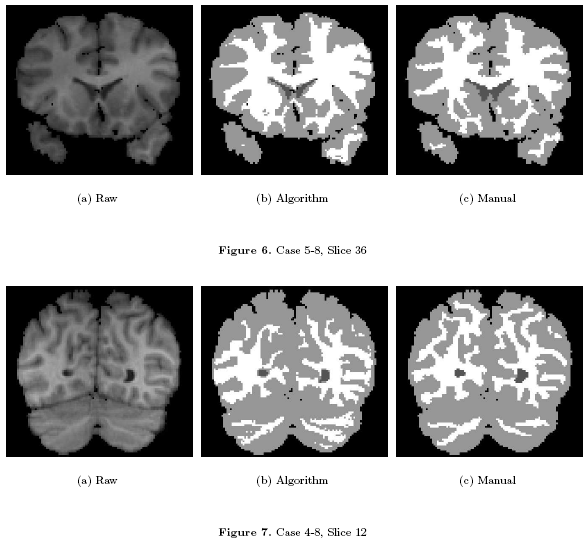
''Some Results'' | ''Some Results'' | ||
| − | * [[Image: | + | * [[Image:Plot white.png | White Matter Performance on the 20 ISBR datasets | 600px]] WM Algorithm Comparisons |
| − | * [[Image: | + | * [[Image:Plot gray.png | Gray Matter Performance on the 20 ISBR datasets | 600px]] GM Algorithm Comparisons |
| − | * [[Image: | + | * [[Image:Fig67.png | Visual Results | 600px]] Visual Results on ISBR data |
''Project Status'' | ''Project Status'' | ||
| Line 41: | Line 55: | ||
* The working ITK code has been committed to the [http://www.na-mic.org:8000/svn/NAMICSandBox/BayesianSegmentationModule/ SandBox] | * The working ITK code has been committed to the [http://www.na-mic.org:8000/svn/NAMICSandBox/BayesianSegmentationModule/ SandBox] | ||
| − | + | = Key Investigators = | |
| − | * | + | * Georgia Tech Algorithms: John Melonakos, Yi Gao, Allen Tannenbaum |
| − | * | + | * Kitware Engineering: Luis Ibanez, Karthik Krishnan |
| − | + | = Publications = | |
| − | * [ | + | ''In Print'' |
| − | + | * [http://www.na-mic.org/publications/pages/display?search=KnowledgeBasedBayesianSegmentation&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&searchbytag=checked&sponsors=checked| NA-MIC Publications Database on Knowledge-Based Bayesian Segmentation] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | [[Category: Segmentation]] [[Category:MRI]] [[Category:Slicer]] | |
| − | |||
Latest revision as of 00:56, 16 November 2013
Home < Projects:KnowledgeBasedBayesianSegmentationBack to NA-MIC Collaborations, Stony Brook University Algorithms, Kitware Engineering
Knowledge Based Bayesian Segmentation
This ITK filter is a segmentation algorithm which utilizes Bayes's Rule along with an affine-invarient anisotropic smoothing filter.
Description
Use Case
I'd like to segment a volume or sub-volume into 'N' classes in a very general manner. I will provide the data and the number of classes that I expect and the algorithm will output a labelmap with 'N' classes.
Data
We have applied this algorithm to 20 normal brain MRI data-sets. We used publicly available data-sets from the Internet Brain Segmentation Repository (IBSR) offered by the Massachusetts General Hospital, Center for Morphometric Analysis. The IBSR data-sets are T1-weighted, 3D coronal brain scans after having been positionally normalized. Manual expert segmentations for these data-sets are publicly available and represent the ground truth used in this work.
Algorithm
This algorithm can be cast in either a static or dynamic framework. In the static framework, the following is the algorithm:
- The user sets the number of distinct classes for segmentation: 'N'
- Generate 'N' prior images (default, 'N' uniform prior images)
- Generate 'N' statistical distributions (default, 'N' normal distributions)
- Generate 'N' membership images by applying the statistical distributions to the raw data
- Generate 'N' posterior images by applying Bayes' rule to the prior and membership images
- Smooth the posterior images for 'm' iterations using an affine-invarient anisotropic smoothing filter and renormalize after each iteration (default, m = 5)
- Apply maximum a posteriori rule to apply labeling and finalize segmentation
In the dynamic framework, the following image depicts the adaptation of the static framework to the dynamic formulation:
The ITK filter design
Some Results
Project Status
- Fully incorporated into itkBayesianClassificationImageFilter and itkBayesianClassificationInitializationImageFilter in ITK CVS.
- Fully wrapped in VTK for use in Slicer.
- The working ITK code has been committed to the SandBox
Key Investigators
- Georgia Tech Algorithms: John Melonakos, Yi Gao, Allen Tannenbaum
- Kitware Engineering: Luis Ibanez, Karthik Krishnan
Publications
In Print