Difference between revisions of "Projects:LabelSpace"
| Line 17: | Line 17: | ||
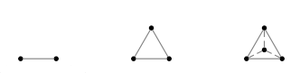
[[Image:Label_space.png|thumb|Figure 1: The first three label space configurations: a unit interval for two labels, a triangle for three labels, and a tetrahedron for four labels (left to right). ]] | [[Image:Label_space.png|thumb|Figure 1: The first three label space configurations: a unit interval for two labels, a triangle for three labels, and a tetrahedron for four labels (left to right). ]] | ||
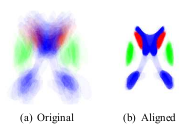
| − | [[Image: | + | [[Image:Ls_original_aligned.png|thumb|Figure 2: Alignment of a set of 30 maps used in the study by Tsai et al. (2003). The original and aligned sets are superimposed for visualization.]] |
= Key Investigators = | = Key Investigators = | ||
Revision as of 17:10, 24 April 2008
Home < Projects:LabelSpaceBack to Georgia Tech Algorithms
Label Space: A Coupled Multi-Shape Representation
Many techniques for multi-shape representation may often develop inaccuracies stemming from either approximations or inherent variation. Label space is an implicit representation that offers unbiased algebraic manipulation and natural expression of label uncertainty. We demonstrate smoothing and registration on multi-label brain MRI.
Description
The surfaces are represented as the zero level set of a signed distance function and shape learning is performed on the embeddings of these shapes. We carry out some experiments to see how well each of these methods can represent a shape, given the training set. We tested the performance of these methods on shapes of left caudate nucleus and left hippocampus. The training set of left caudate nucleus consisted of 26 data sets and the test set contained 3 volumes. Error between a particular shape representation and ground truth was calculated by computing the number of mislabeled voxels using each of the methods. Figure 1 gives the error using each of the methods. Similar tests were done on a training set of 20 hippocampus data with 3 test volumes. Figure 2 gives the error table for each of the methods [1].
Key Investigators
- Georgia Tech Algorithms: James Malcolm, Yogesh Rathi, Allen Tannenbaum
Publications
J. Malcolm, Y. Rathi, and A. Tannenbaum. "Label Space: A Multi-Object Shape Representation." In Combinatorial Image Analysis, 2008.