Difference between revisions of "Projects:MGH-HeadAndNeck-PtSetReg"
Ivan.kolesov (talk | contribs) (Created page with ' Back to Boston University Algorithms __NOTOC__ = Interactive Image Segmentation With Active Contours = A driving clinical study for the present work is a popul…') |
|||
| (13 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[Algorithm: | + | Back to [[Algorithm:Stony Brook|Stony Brook University Algorithms]] |
__NOTOC__ | __NOTOC__ | ||
| − | = | + | = Semi-Automatic Image Registration = |
| − | + | We recognize that the difference between a failure of an automatic image registration approach and a success of a semi-automatic method can be a small amount of user input. The goal of this work is to register two CT volumes of different patients that are related by a large misalignment. The user sets two thresholds for each image: one for the bone mask and another for the flesh tissue. This operation is not time consuming but simplifies the registration task dramatically for the automatic algorithm. | |
= Description = | = Description = | ||
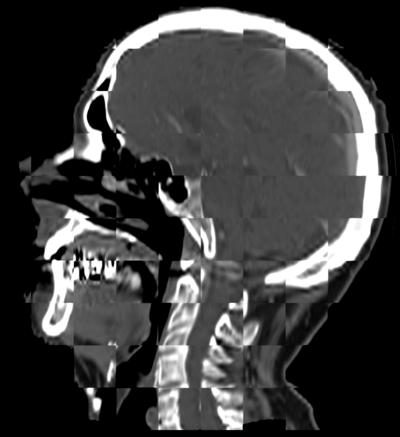
| − | In this | + | In this example, large misalignment is present between the two patients. |
| + | * [[Image:PreRegFleshSkeleton.png | PreRegFleshSkeleton| 400px]] | ||
| + | Original Misalignment of the volumes. | ||
| + | |||
| + | Point clouds are generated from label maps of bone. The computed registration field, which is guaranteed to be injective, is applied to the original CT volumes. | ||
| + | |||
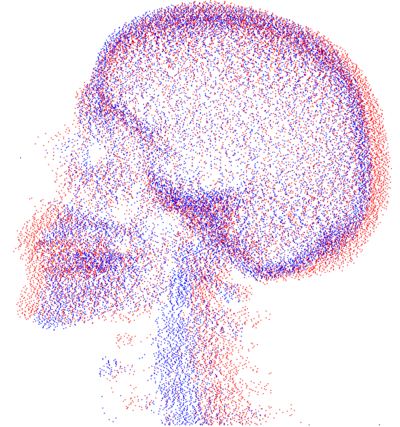
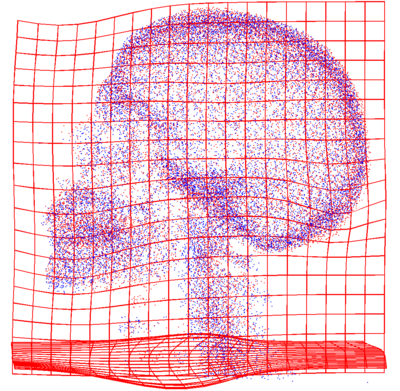
| + | * [[Image:SkeletonMisalignedView1.png | SkeletonMisalignedView1| 400px]] [[Image:SkeletonAlignedView1.png | SkeletonMisalignedView1| 400px]] | ||
| + | Point clouds representing bone tissue of the patients (before and after registration). | ||
| + | |||
| + | |||
| + | Another set of point clouds is generated by sampling from label maps of flesh. To avoid undoing the previous registration, regions belonging to the registered bone tissue from above are constrained not to move. Again, an injective deformation field is computed. | ||
| + | |||
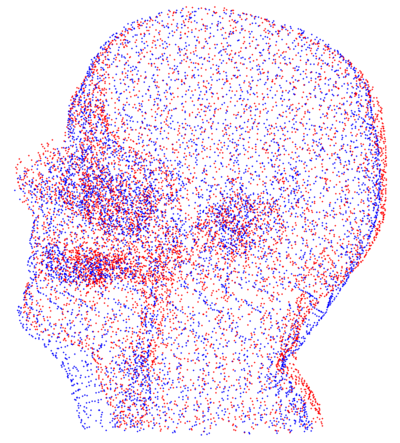
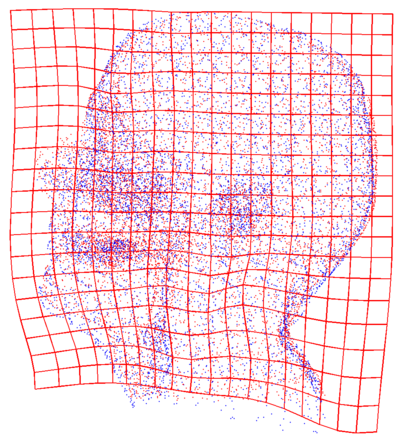
| + | * [[Image:FleshMis.png | FleshMis| 400px]] [[Image:FleshAlignedView1.png | FleshAlignedView1| 400px]] | ||
| + | Point clouds representing flesh tissue of the patients (before and after registration). This step is constrained. | ||
| + | |||
| + | The result of applying the two deformations computed by the proposed process are shown below. | ||
| + | |||
| + | * [[Image:PostRegFleshSkeleton.png | PostRegFleshSkeleton| 400px]] | ||
| + | Aligned images using the two step registration process. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Current State of Work == | == Current State of Work == | ||
| − | + | A pipeline composed of Matlab and mex-ed C++ code has been implemented. | |
= Key Investigators = | = Key Investigators = | ||
| − | * Georgia Tech: Ivan Kolesov, | + | * Georgia Tech: Ivan Kolesov, Patricio Vela |
| − | * Boston University: Allen Tannenbaum | + | * Boston University: Jehoon Lee, Allen Tannenbaum |
| − | * | + | * MGH: Gregory Sharp |
= Publications = | = Publications = | ||
| Line 30: | Line 42: | ||
''In Press'' | ''In Press'' | ||
| − | I. Kolesov | + | I. Kolesov, J. Lee, P.Vela, G. Sharp and A. Tannenbaum. Diffeomorphic Point Set Registration with Landmark Constraints. In Preparation for PAMI. |
| − | |||
| − | |||
| − | P. | ||
Latest revision as of 01:03, 16 November 2013
Home < Projects:MGH-HeadAndNeck-PtSetRegBack to Stony Brook University Algorithms
Semi-Automatic Image Registration
We recognize that the difference between a failure of an automatic image registration approach and a success of a semi-automatic method can be a small amount of user input. The goal of this work is to register two CT volumes of different patients that are related by a large misalignment. The user sets two thresholds for each image: one for the bone mask and another for the flesh tissue. This operation is not time consuming but simplifies the registration task dramatically for the automatic algorithm.
Description
In this example, large misalignment is present between the two patients.
Original Misalignment of the volumes.
Point clouds are generated from label maps of bone. The computed registration field, which is guaranteed to be injective, is applied to the original CT volumes.
Point clouds representing bone tissue of the patients (before and after registration).
Another set of point clouds is generated by sampling from label maps of flesh. To avoid undoing the previous registration, regions belonging to the registered bone tissue from above are constrained not to move. Again, an injective deformation field is computed.
Point clouds representing flesh tissue of the patients (before and after registration). This step is constrained.
The result of applying the two deformations computed by the proposed process are shown below.
Aligned images using the two step registration process.
Current State of Work
A pipeline composed of Matlab and mex-ed C++ code has been implemented.
Key Investigators
- Georgia Tech: Ivan Kolesov, Patricio Vela
- Boston University: Jehoon Lee, Allen Tannenbaum
- MGH: Gregory Sharp
Publications
In Press
I. Kolesov, J. Lee, P.Vela, G. Sharp and A. Tannenbaum. Diffeomorphic Point Set Registration with Landmark Constraints. In Preparation for PAMI.