Difference between revisions of "Projects:MGH-HeadAndNeck-RT"
Ivan.kolesov (talk | contribs) |
|||
| (16 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | = | + | Back to [[Algorithm:Stony Brook|Stony Brook University Algorithms]] |
| + | __NOTOC__ | ||
| + | = Adaptive Radiotherapy for head, neck and thorax = | ||
| − | Proton therapy is used to deliver accurate doses of radiation to people undergoing cancer treatment. At the beginning of treatment, a personalized plan describing the amount of radiation and location to which it must be delivered is created for the patient. However, over the course of the treatment, which lasts weeks, a person's anatomy is likely to change. Adaptive radiotherapy aims to improve fractionated radiotherapy by re-optimizing the radiation treatment plan for each session. To update the plan, the CT images acquired during a treatment session are registered to the treatment plan and doses of radiation to be delivered are re-calculated accordingly. | + | Proton therapy is used to deliver accurate doses of radiation to people undergoing cancer treatment. At the beginning of treatment, a personalized plan describing the amount of radiation and location to which it must be delivered is created for the patient. However, over the course of the treatment, which lasts weeks, a person's anatomy is likely to change. Adaptive radiotherapy aims to improve fractionated radiotherapy by re-optimizing the radiation treatment plan for each session. To update the plan, the CT images acquired during a treatment session are registered to the treatment plan and doses of radiation to be delivered are re-calculated accordingly. Registering a patient scan to a model (ex.Atlas) provides important prior information for a segmentation algorithm and in the other direction, having segmentation of a structure, can be used for better registration; hence, segmentation and registration must be done concurrently. |
| − | ==Segmentation | + | = Description = |
| + | |||
| + | Initial experiments show that bony structures such as the mandible can be segmented accurately with a variational active contour. However, for soft tissue such as the brain stem, the intensity profile does not contain sufficient information for reasonably accurate segmentation. To deal with "soft boundaries" infinite dimensional active contours must be constrained by using shape priors and/or interactive user input. One way to constrain a segmentation is shown in our work in MTNS; there, known spatial relationships between structures is exploited. First, a structure we are confident in will be segmented. Using probabilistic PCA a metric used to describe how likely the structure whose segmentation we have obtained is; this metric is essentially a description of how confident we are in the correct segmentation. Then, the location of this structure will be used as prior information(it becomes a landmark) to segment a more difficult structure. Iteratively, the nth structure to be segmented will have n-1 priors do draw information from with a confidence metric for each prior. The likely location of the nth structure, calculated as described above, will serve as an input to constrain an active contours algorithm. Additionally, we have had excellent results when constraining structures of the eye to simple geometrical shapes such as ellipses and tubular to limit the number of free parameter. A sample segmentation of the eye ball is shown below. | ||
| + | |||
| + | * [[Image:Model3D_upTrans.png | Rel Pred| 250px]] Prediction of mandible location(red) give a brainstem(yellow) and larynx(blue) segmentation; ground truth segmenation is green. | ||
| + | * [[Image:3D_eye.png | Eye Seg| 250px]] Segmentation of the eye ball. | ||
| − | |||
== Current State of Work == | == Current State of Work == | ||
| + | We have gathered a sample data set; currently, we are registering the scan to create a united patient model. This model will localize structures and constrain shape. Additionally, we are investigation interaction approaches for registration and segmentation that will "put the user in the loop" but greatly reduce the work compared to manual approaches. | ||
| + | |||
| − | + | = Key Investigators = | |
| − | + | * Georgia Tech: Ivan Kolesov, Vandana Mohan, and Allen Tannenbaum | |
| + | * Massachusetts General Hospital: Gregory Sharp | ||
| − | + | = Publications = | |
| − | + | ''In Press'' | |
| − | + | I. Kolesov, V. Mohan, G. Sharp and A. Tannenbaum. Coupled Segmentation for Anatomical Structures by Combining Shape and Relational Spatial Information. MTNS 2010. | |
| − | |||
Latest revision as of 01:02, 16 November 2013
Home < Projects:MGH-HeadAndNeck-RTBack to Stony Brook University Algorithms
Adaptive Radiotherapy for head, neck and thorax
Proton therapy is used to deliver accurate doses of radiation to people undergoing cancer treatment. At the beginning of treatment, a personalized plan describing the amount of radiation and location to which it must be delivered is created for the patient. However, over the course of the treatment, which lasts weeks, a person's anatomy is likely to change. Adaptive radiotherapy aims to improve fractionated radiotherapy by re-optimizing the radiation treatment plan for each session. To update the plan, the CT images acquired during a treatment session are registered to the treatment plan and doses of radiation to be delivered are re-calculated accordingly. Registering a patient scan to a model (ex.Atlas) provides important prior information for a segmentation algorithm and in the other direction, having segmentation of a structure, can be used for better registration; hence, segmentation and registration must be done concurrently.
Description
Initial experiments show that bony structures such as the mandible can be segmented accurately with a variational active contour. However, for soft tissue such as the brain stem, the intensity profile does not contain sufficient information for reasonably accurate segmentation. To deal with "soft boundaries" infinite dimensional active contours must be constrained by using shape priors and/or interactive user input. One way to constrain a segmentation is shown in our work in MTNS; there, known spatial relationships between structures is exploited. First, a structure we are confident in will be segmented. Using probabilistic PCA a metric used to describe how likely the structure whose segmentation we have obtained is; this metric is essentially a description of how confident we are in the correct segmentation. Then, the location of this structure will be used as prior information(it becomes a landmark) to segment a more difficult structure. Iteratively, the nth structure to be segmented will have n-1 priors do draw information from with a confidence metric for each prior. The likely location of the nth structure, calculated as described above, will serve as an input to constrain an active contours algorithm. Additionally, we have had excellent results when constraining structures of the eye to simple geometrical shapes such as ellipses and tubular to limit the number of free parameter. A sample segmentation of the eye ball is shown below.
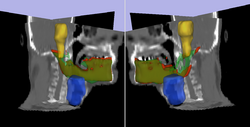 Prediction of mandible location(red) give a brainstem(yellow) and larynx(blue) segmentation; ground truth segmenation is green.
Prediction of mandible location(red) give a brainstem(yellow) and larynx(blue) segmentation; ground truth segmenation is green. Segmentation of the eye ball.
Segmentation of the eye ball.
Current State of Work
We have gathered a sample data set; currently, we are registering the scan to create a united patient model. This model will localize structures and constrain shape. Additionally, we are investigation interaction approaches for registration and segmentation that will "put the user in the loop" but greatly reduce the work compared to manual approaches.
Key Investigators
- Georgia Tech: Ivan Kolesov, Vandana Mohan, and Allen Tannenbaum
- Massachusetts General Hospital: Gregory Sharp
Publications
In Press
I. Kolesov, V. Mohan, G. Sharp and A. Tannenbaum. Coupled Segmentation for Anatomical Structures by Combining Shape and Relational Spatial Information. MTNS 2010.