Difference between revisions of "Projects:MethodEvaluationValidation"
| Line 20: | Line 20: | ||
= Publications = | = Publications = | ||
''In Print'' | ''In Print'' | ||
| − | * [http://www.na-mic.org/publications/pages/display?search=MethodEvaluationValidation&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&sponsors=checked&searchbytag=checked| NA-MIC Publication Database] | + | * [http://www.na-mic.org/publications/pages/display?search=MethodEvaluationValidation&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&sponsors=checked&searchbytag=checked| NA-MIC Publication Database on Evaluation and Comparison of Medical Image Analysis Methods] |
= Links = | = Links = | ||
Revision as of 20:24, 11 May 2010
Home < Projects:MethodEvaluationValidationBack to UNC Algorithms
Evaluation and Comparison of Medical Image Analysis Methods
In this project, we want to focus on the evaluation of medical image analysis methods for specific clinical applications in respect to development of evaluation methodology and the organization of venues promoting such comparison and validation studies.
Description
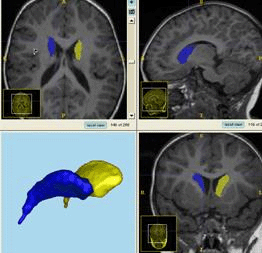
We have developed an open source tool for the evaluation of 3D segmentation algorithms combining a variety of segmentation comparison methods with respect to the performance of human expert raters. This tool was employed during the workshop on "3D Segmentation in the Clinic - A Grand Challenge", on October 26, 2007 held in conjunction with MICCAI 2007 and continues to be used in workshop attached websites on caudate segmentation (cause07.org) and liver segmentation (sliver07.org). On these webpages, you can browse the results of various systems, and read the respective papers and descriptions about the systems. Anybody can join the competition by registering a team, downloading the training and test datasets, and submitting the segmentation results. The MICCAI 2007 workshop was organized in collaboration with Tobias Heimann (German Cancer Research Center, Liver Segmentation) and Bram van Ginneken (University of Utrecht, general organization & webpage development). There were close to 70 participants at the workshop with an overwhelmingly positive feedback.
A version adapted for lesion segmentation (in light of the MIND DBP 2 application) was developed for the 2008 MICCAI workshop with special focus on lesion detection metrics. The workshop consisted on 3 challenges: Multiple Sclerosis lesion segmentation, Liver tumor segmentation and artery central lumen line extraction. Next to our team, the workshop was co-organized with the Children Hospital of Boston, Siemens Medical Systems and Erasmus University (Rotterdam, Netherlands). As in the previous year, the workshop was a full success and with over 70 participants MICCAI's largest workshop this year. The MS lesion segmentation is continued online. The 2008 workshop proceedings were also publicly released on the online published MIDAS journal.
In 2009, the third installment of the workshop took place. We provided significant advice to this year's organizers and did not directly organize one of the challenges. The success also in 2009 reassures us though of the continued need for such evaluation workshops and we are again directly involved for the 2010 MICCAI workshop. The challenges this year encompass knee bone and cartilage segmentation, parotid gland segmentation and lung CT registration.
Key Investigators
- UNC: Martin Styner
Publications
In Print