Difference between revisions of "Projects:MultiScaleShapeSegmentation"
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[Algorithm: | + | Back to [[Algorithm:Stony Brook|Stony Brook University Algorithms]] |
__NOTOC__ | __NOTOC__ | ||
= Multi-scale Shape Representation and Segmentation With Applications to Radiotherapy = | = Multi-scale Shape Representation and Segmentation With Applications to Radiotherapy = | ||
= Description = | = Description = | ||
| − | + | Brain metastases constitute the most common manifestation of cancer involving the central nervous system. Indeed, it has been estimated that over 170,000 cases of brain metastases are diagnosed annually in the United States alone [1]. During the past half-century, the cornerstone of treatment for this oncologic phenomenon has been whole brain irradiation (WBI) [2]. WBI has multiple salutary effects including rapid relief of neurological signs and symptoms as well as enhanced local control. Moreover, WBI represents an attractive clinical alternative because it can potentially suppress micrometastases that are undetectable with the current degree of resolution by MR imaging. Unfortunately, WBI may also engender side effects including memory deficits and decrements in quality of life [3]. | |
| + | |||
| + | Stereotactic irradiation is an option that has gained popularity in the management of brain metastases. Stereotactic irradiation is appealing because it is of short duration, uses multiple intersecting beams to augment the dose within the tumor volume and provides a rapid dose fall-off thereby optimizing the dosimetric gradient outside the tumor. This rationale allowed Chang et al. [4] to mount a trial comparing WBI to stereotactic techniques for patients suffering from brain metastases. The results of the trial indicated less decline in learning and memory function within the stereotactic arm. Yet, others have questioned whether an unacceptable subset of patients (among those treated focally) failed intracranially, albeit outside the radiosurgical treatment volumes. | ||
| + | |||
| + | In order for those receiving WBI to obtain the optimal intracranial control of disease while simultaneously preserving neurocognitive function, it is of strategic importance to recognize that the primary neurocognitive impairments are in memory. Since memory control is thought to be mediated by the hippocampus, attention has been turned to whole brain radiotherapeutic techniques that allow the sparing of the hippocampus. In order to be able to minimize dose deposition within the hippocampus, clinicians must be able to confidently identify that structure. The accuracy and consistency of segmentation can be improved by automating the process and including shape prior knowledge. Also, segmentation is a necessary step prior to registration; segmented structures provide landmarks and can be used to limit the number of free variables in deformable registration, which, in turn, leads to more accurate results. | ||
| + | |||
| + | In order to automatically extract the hippocampus region from the MR images, we present in this work a multiscale representation for shapes with arbitrary topology, and a method to segment the target organ/tissue from medical images having very low contrast with respect to surrounding regions using multiscale shape information and local image features. In many previous works, shape knowledge was incorporated by first constructing a shape space from training cases, and then constraining the segmentation process to be within the learned shape space. However, such an approach has certain limitations due to the number of variations in the learned shape space. Moreover, small scale shape variances are usually overwhelmed by those in the large scale, and therefore the local shape information is lost. In this work, first we handle this problem by providing a multiscale shape representation using the wavelet transform. Consequently, the shape variances captured by the statistical learning step are also represented at various scales. In doing so, not only the number of shape variances are largely enriched, but also the small scale changes are nicely captured. Furthermore, in order to make full use of the training information, not only the shape but also the grayscale training images are utilized in a multi-atlas initialization procedure. | ||
== Result == | == Result == | ||
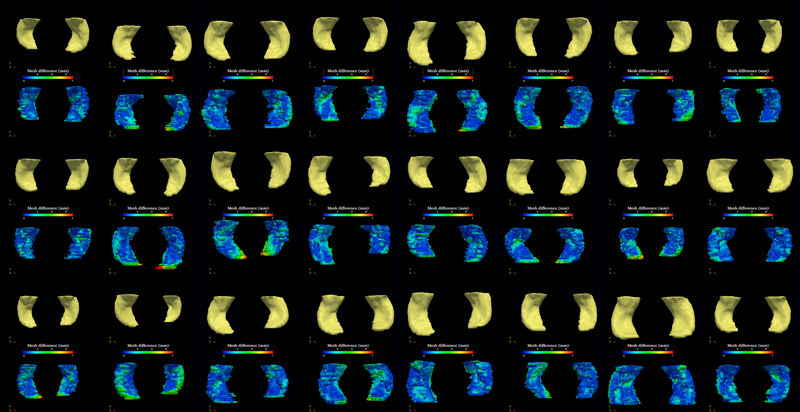
| − | The method is applied on hippocampus. | + | The segmentation method is applied on hippocampus. In the figure below, we show the segmentation results. In the first, third, and the fifth rows, the yellow colored shapes are the segmentation results output by the method. In the second, forth, and sixth rows, the colors on the shapes indicate the difference with the manual segmentation results: For each point on the shape (result of the segmentation algorithm), we compute the closest point on the manual segmented surface, and record the distance to that point. Such distances are encoded by the color shown in those rows. |
[[Image:MultiScaleHippoSegmentationHausdorf.png|800px]] | [[Image:MultiScaleHippoSegmentationHausdorf.png|800px]] | ||
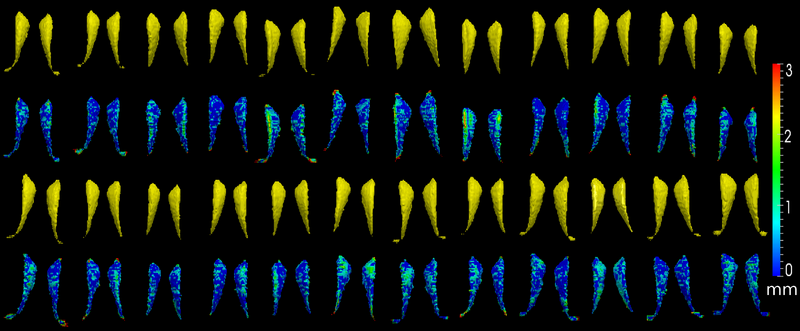
| − | The method is also applied on caudate | + | The method is also applied on caudate. Similarly to the hippocampus cases, in the figure below, we show the segmentation results. In the first and third rows, the yellow colored shapes are the segmentation results output by the method. In the second and forth rows, the colors on the shapes indicate the difference with the manual segmentation results: For each point on the shape (result of the segmentation algorithm), we compute the closest point on the manual segmented surface, and record the distance to that point. Such distances are encoded by the color shown in those rows. |
[[Image:MultiScaleCaudateSegmentationHausdorf.png|800px]] | [[Image:MultiScaleCaudateSegmentationHausdorf.png|800px]] | ||
| Line 20: | Line 26: | ||
Georgia Tech: Yi Gao and Allen Tannenbaum | Georgia Tech: Yi Gao and Allen Tannenbaum | ||
| + | |||
| + | = References = | ||
| + | # Gao, Y. and Corn, B. and Schifter, D. and Tannenbaum, A. ''Multiscale 3D Shape Representation and Segmentation with Applications to Hippocampal/Caudate Extraction from Brain MRI'', Medical Image Analysis, 2011 in press | ||
| + | # Patchell, R.: The management of brain metastases. Cancer Treat Rev. 29(6), 533–540 (2003) | ||
| + | # Knisely, J.: Focused attention on brain metastases. Lancet Oncol. 10, 1037–1044 (2009) | ||
| + | # Aoyama, H., Tago, M., Kato, N., et al.: Neurocognitive function of patients with brain metastasis who received either whole brain radiotherapy plus stereotactic radiosurgery or radiosurgery alone. In: Int. J. Radiat Oncol, vol. 68(5), pp. 1388–1395 (2007) | ||
| + | # Chang, E., Wefel, J., Hess, K., et al.: Neurocognition in patients with brain metastases treated with radiosurgery or radiosurgery plus whole-brain irradiation: a randomised controlled trial. Lancet Oncology 10(11), 1037–1044 (2009) | ||
Latest revision as of 01:03, 16 November 2013
Home < Projects:MultiScaleShapeSegmentationBack to Stony Brook University Algorithms
Multi-scale Shape Representation and Segmentation With Applications to Radiotherapy
Description
Brain metastases constitute the most common manifestation of cancer involving the central nervous system. Indeed, it has been estimated that over 170,000 cases of brain metastases are diagnosed annually in the United States alone [1]. During the past half-century, the cornerstone of treatment for this oncologic phenomenon has been whole brain irradiation (WBI) [2]. WBI has multiple salutary effects including rapid relief of neurological signs and symptoms as well as enhanced local control. Moreover, WBI represents an attractive clinical alternative because it can potentially suppress micrometastases that are undetectable with the current degree of resolution by MR imaging. Unfortunately, WBI may also engender side effects including memory deficits and decrements in quality of life [3].
Stereotactic irradiation is an option that has gained popularity in the management of brain metastases. Stereotactic irradiation is appealing because it is of short duration, uses multiple intersecting beams to augment the dose within the tumor volume and provides a rapid dose fall-off thereby optimizing the dosimetric gradient outside the tumor. This rationale allowed Chang et al. [4] to mount a trial comparing WBI to stereotactic techniques for patients suffering from brain metastases. The results of the trial indicated less decline in learning and memory function within the stereotactic arm. Yet, others have questioned whether an unacceptable subset of patients (among those treated focally) failed intracranially, albeit outside the radiosurgical treatment volumes.
In order for those receiving WBI to obtain the optimal intracranial control of disease while simultaneously preserving neurocognitive function, it is of strategic importance to recognize that the primary neurocognitive impairments are in memory. Since memory control is thought to be mediated by the hippocampus, attention has been turned to whole brain radiotherapeutic techniques that allow the sparing of the hippocampus. In order to be able to minimize dose deposition within the hippocampus, clinicians must be able to confidently identify that structure. The accuracy and consistency of segmentation can be improved by automating the process and including shape prior knowledge. Also, segmentation is a necessary step prior to registration; segmented structures provide landmarks and can be used to limit the number of free variables in deformable registration, which, in turn, leads to more accurate results.
In order to automatically extract the hippocampus region from the MR images, we present in this work a multiscale representation for shapes with arbitrary topology, and a method to segment the target organ/tissue from medical images having very low contrast with respect to surrounding regions using multiscale shape information and local image features. In many previous works, shape knowledge was incorporated by first constructing a shape space from training cases, and then constraining the segmentation process to be within the learned shape space. However, such an approach has certain limitations due to the number of variations in the learned shape space. Moreover, small scale shape variances are usually overwhelmed by those in the large scale, and therefore the local shape information is lost. In this work, first we handle this problem by providing a multiscale shape representation using the wavelet transform. Consequently, the shape variances captured by the statistical learning step are also represented at various scales. In doing so, not only the number of shape variances are largely enriched, but also the small scale changes are nicely captured. Furthermore, in order to make full use of the training information, not only the shape but also the grayscale training images are utilized in a multi-atlas initialization procedure.
Result
The segmentation method is applied on hippocampus. In the figure below, we show the segmentation results. In the first, third, and the fifth rows, the yellow colored shapes are the segmentation results output by the method. In the second, forth, and sixth rows, the colors on the shapes indicate the difference with the manual segmentation results: For each point on the shape (result of the segmentation algorithm), we compute the closest point on the manual segmented surface, and record the distance to that point. Such distances are encoded by the color shown in those rows.
The method is also applied on caudate. Similarly to the hippocampus cases, in the figure below, we show the segmentation results. In the first and third rows, the yellow colored shapes are the segmentation results output by the method. In the second and forth rows, the colors on the shapes indicate the difference with the manual segmentation results: For each point on the shape (result of the segmentation algorithm), we compute the closest point on the manual segmented surface, and record the distance to that point. Such distances are encoded by the color shown in those rows.
Key Investigators
Georgia Tech: Yi Gao and Allen Tannenbaum
References
- Gao, Y. and Corn, B. and Schifter, D. and Tannenbaum, A. Multiscale 3D Shape Representation and Segmentation with Applications to Hippocampal/Caudate Extraction from Brain MRI, Medical Image Analysis, 2011 in press
- Patchell, R.: The management of brain metastases. Cancer Treat Rev. 29(6), 533–540 (2003)
- Knisely, J.: Focused attention on brain metastases. Lancet Oncol. 10, 1037–1044 (2009)
- Aoyama, H., Tago, M., Kato, N., et al.: Neurocognitive function of patients with brain metastasis who received either whole brain radiotherapy plus stereotactic radiosurgery or radiosurgery alone. In: Int. J. Radiat Oncol, vol. 68(5), pp. 1388–1395 (2007)
- Chang, E., Wefel, J., Hess, K., et al.: Neurocognition in patients with brain metastases treated with radiosurgery or radiosurgery plus whole-brain irradiation: a randomised controlled trial. Lancet Oncology 10(11), 1037–1044 (2009)