Projects:MultiScaleShapeSegmentation
Back to Georgia Tech Algorithms
Multi-scale Shape Representation and Segmentation With Applications to Radiotherapy
Description
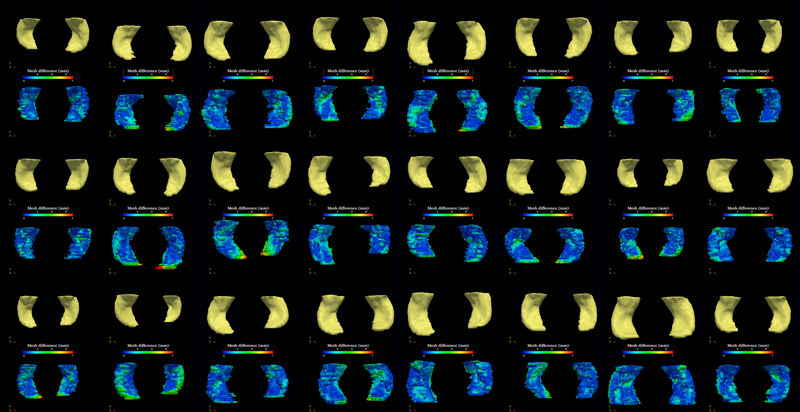
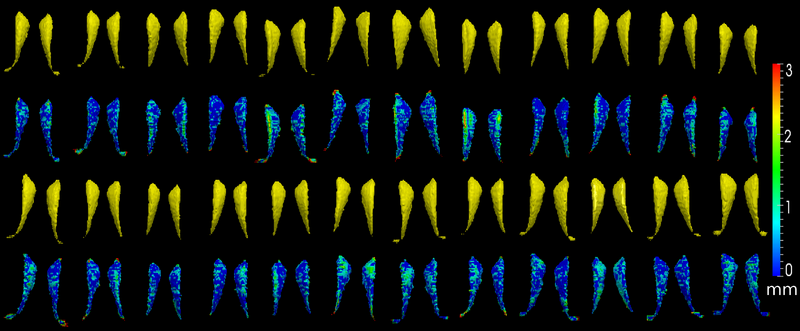
We present in this work a multiscale representation for shapes with arbitrary topology, and a method to segment the target organ/tissue from medical images having very low contrast with respect to surrounding regions using multiscale shape information and local image features. In many previous works, shape knowledge was incorporated by first constructing a shape space from training cases, and then constraining the segmentation process to be within the learned shape space. However, such an approach has certain limitations due to the number of variations in the learned shape space. Moreover, small scale shape variances are usually overwhelmed by those in the large scale, and therefore the local shape information is lost. In this work, first we handle this problem by providing a multiscale shape representation using the wavelet transform. Consequently, the shape variances captured by the statistical learning step are also represented at various scales. In doing so, not only the number of shape variances are largely enriched, but also the small scale changes are nicely captured. Furthermore, in order to make full use of the training information, not only the shape but also the grayscale training images are utilized in a multi-atlas initialization procedure.
Result
The method is applied on hippocampus.
The method is also applied on caudate
Key Investigators
Georgia Tech: Yi Gao and Allen Tannenbaum