Difference between revisions of "Projects:MultiscaleShapeSegmentation"
| (25 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm:Stony Brook|Stony Brook University Algorithms]], [[Algorithm:UNC|UNC Algorithms]], [[Engineering:GE|GE Engineering]], [[Engineering:Kitware|Kitware Engineering]], [[DBP1:Harvard|Harvard DBP 1]] | ||
| + | __NOTOC__ | ||
= Multiscale Shape Segmentation = | = Multiscale Shape Segmentation = | ||
| − | + | To represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. | |
| − | + | = Description = | |
| − | |||
| − | |||
| − | + | == Shape Representation and Prior == | |
| − | |||
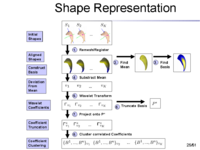
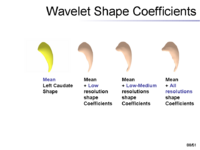
The overview of our shape representation is given in Figure 1. Our technique defines a multiscale parametric model of surfaces belonging to the same population using a compact set of spherical wavelets targeted to that population (Figure 2). We further refine the shape representation by separating into groups wavelet coefficients that describe independent global and/or local biological variations in the population, using spectral graph partitioning. We then learn a prior probability distribution induced over each group to explicitly encode these variations at different scales and spatial locations (Figure 4) [1]. | The overview of our shape representation is given in Figure 1. Our technique defines a multiscale parametric model of surfaces belonging to the same population using a compact set of spherical wavelets targeted to that population (Figure 2). We further refine the shape representation by separating into groups wavelet coefficients that describe independent global and/or local biological variations in the population, using spectral graph partitioning. We then learn a prior probability distribution induced over each group to explicitly encode these variations at different scales and spatial locations (Figure 4) [1]. | ||
| Line 15: | Line 14: | ||
[[Image:Gatech_SW_mscale_shape.png|thumb|200px|Figure 2: A shape is represented using spherical wavelet coefficients]] | [[Image:Gatech_SW_mscale_shape.png|thumb|200px|Figure 2: A shape is represented using spherical wavelet coefficients]] | ||
| − | + | == Segmentation == | |
| − | |||
| − | + | Based on this representation, we derive a parametric active surfaceIn evolution using the multiscale prior coefficients as parameters for our optimization procedure to naturally include the prior for segmentation. Additionally, the optimization method can be applied in a coarse-to-fine manner. | |
| − | |||
| − | + | == Results == | |
| − | |||
| − | |||
| − | + | We applied our algorithm to the caudate nucleus, a brain structure of interest in the study of schizophrenia [2]. Our validation shows our algorithm is computationally efficient and outperforms the Active Shape Model (ASM) algorithm, by capturing finer shape details. | |
| − | |||
| − | |||
| − | |||
| − | + | = Key Investigators = | |
| − | * | + | * Georgia Tech Algorithms: Delphine Nain, Aaron Bobick, Allen Tannenbaum |
| − | * | + | * UNC Algorithms: Martin Styner |
| − | * | + | * GE Engineering: Jim Miller |
| + | * Kitware Engineering: Luis Ibanez | ||
| + | * Harvard DBP 1: Steven Haker, James Levitt, Marc Niethammer, Sylvain Bouix, Martha Shenton | ||
| − | + | = Publications = | |
| − | * | + | ''In Print'' |
| − | + | * [http://www.na-mic.org/publications/pages/display?search=MultiscaleShapeSegmentation&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&searchbytag=checked&sponsors=checked| NA-MIC Publications Database on Multiscale Shape Segmentation Techniques] | |
| − | |||
| − | |||
| − | [[Category: | + | [[Category:Shape Analysis]] [[Category:Segmentation]] [[Category:MRI]] [[Category:Schizophrenia]] |
Latest revision as of 00:58, 16 November 2013
Home < Projects:MultiscaleShapeSegmentationBack to NA-MIC Collaborations, Stony Brook University Algorithms, UNC Algorithms, GE Engineering, Kitware Engineering, Harvard DBP 1
Multiscale Shape Segmentation
To represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus.
Description
Shape Representation and Prior
The overview of our shape representation is given in Figure 1. Our technique defines a multiscale parametric model of surfaces belonging to the same population using a compact set of spherical wavelets targeted to that population (Figure 2). We further refine the shape representation by separating into groups wavelet coefficients that describe independent global and/or local biological variations in the population, using spectral graph partitioning. We then learn a prior probability distribution induced over each group to explicitly encode these variations at different scales and spatial locations (Figure 4) [1].
Segmentation
Based on this representation, we derive a parametric active surfaceIn evolution using the multiscale prior coefficients as parameters for our optimization procedure to naturally include the prior for segmentation. Additionally, the optimization method can be applied in a coarse-to-fine manner.
Results
We applied our algorithm to the caudate nucleus, a brain structure of interest in the study of schizophrenia [2]. Our validation shows our algorithm is computationally efficient and outperforms the Active Shape Model (ASM) algorithm, by capturing finer shape details.
Key Investigators
- Georgia Tech Algorithms: Delphine Nain, Aaron Bobick, Allen Tannenbaum
- UNC Algorithms: Martin Styner
- GE Engineering: Jim Miller
- Kitware Engineering: Luis Ibanez
- Harvard DBP 1: Steven Haker, James Levitt, Marc Niethammer, Sylvain Bouix, Martha Shenton
Publications
In Print