Projects:ParticlesForShapesAndComplexes
Back to NA-MIC Collaborations, Utah Algorithms
Shape Modeling and Analysis with Particle Systems
Overview
This work addresses technical challenges in biomedical shape analysis through the development of novel modeling and analysis methodologies, and seeks validation of those methodologies by their application to real-world research problems. The main focus of the work is the development and validation of a new framework for constructing compact statistical point-based models of ensembles of similar shapes that does not rely on any specific surface parameterization. The proposed optimization uses an entropy-based minimization that balances the simplicity of the model (compactness) with the accuracy of the surface representations. This framework is easily extended to handle more general classes of shape modeling, such as multiple-object complexes and correspondence based on functions of position. This work also addresses the issue of how to do hypothesis testing with the proposed modeling framework, since, to date, the shape analysis community has not reached a consensus regarding a systematic approach to statistical analysis with point-based models. Finally, another important issue that remains is how to visualize significant shape differences in a way that allows researchers to understand not only whether differences exist, but what those shape differences are. This latter consideration is obviously of importance in in relating shape differences to scientific hypotheses.
The following list is a summary of research and development results to date.
- We have implemented a mathematical framework and a robust numerical algorithm implementation for computing optimized correspondence-point shape models using an entropy-based optimization and particle-system technology.
- We have proposed a general methodology for hypothesis testing with point-based shape models that is suitable for use with the particle-based correspondence algorithm. Additionally, we have proposed ideas for visualization to aid in the interpretation of these shape statistics.
- The particle-based correspondence algorithm and statistical analysis methodology have been extended to more general classes of shape analysis problems: (a) the analysis of multiple-object complexes and (b) the generalization to correspondences based on generic functions of position.
- In cooperation with scientists and clinicians, we have published several papers that evaluate the above methodologies in the context of biomedical research.
Section 1: Particle-Based Correspondence Optimization
The proposed correspondence optimization method is to construct a point-based sampling of a shape ensemble that simultaneously maximizes both the geometric accuracy and the statistical simplicity of the model. Surface point samples, which also define the shape-to-shape correspondences, are modeled as sets of dynamic particles that are constrained to lie on a set of implicit surfaces. Sample positions are optimized by gradient descent on an energy function that balances the negative entropy of the distribution on each shape with the positive entropy of the ensemble of shapes. We have also extended the method with a curvature-adaptive sampling strategy in order to better approximate the geometry of the objects.
Any set of implicitly defined surfaces, such as a set of binary segmentations, is appropriate as input to this framework. A binary mask, for example, contains an implicit shape surface at the interface of the labeled pixels and the background. Because the proposed method is completely generalizable to higher dimensions, we are able to process shapes in 2D and 3D using the same C++ software implementation, which is templated by dimension. Processing time on a 2GHz desktop machine scales linearly with the number of particles in the system and ranges from 20 minutes for a 2D system of a few thousand particles to several hours for a 3D system of tens of thousands of particles.
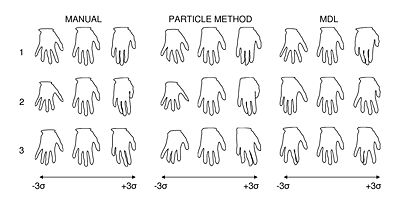
We have found that the proposed particle method compares favorably to state-of-the-art methods for optimizing surface point correspondences. Figure 1, for example, illustrates an experiment on a set hand shape contours that shows a favorable comparison with the Minimum Description Length (MDL) method of Davies, et al. [R Davies, et al., IEEE Trans Med Imaging, V. 21, No. 5, 2002]. In the figure, we also compare our results with a set of ideal, manually selected correspondences, which introduce anatomical knowledge of the digits. Principal component analysis shows that the proposed particle method and the manually selected points both produce very similar models, while the MDL model differs significantly.
Existing 3D MDL implementations rely on spherical parameterizations, and are therefore only capable of analyzing shapes topologically equivalent to a sphere. The particle-based method does not have this limitation. As a demonstration of this, we applied the proposed method to a set of 25 randomly chosen tori, drawn from a 2D, normal distribution parameterized by the small radius r and the large radius R. An analysis of the resulting model showed that the particle system method discovered the two pure modes of variation, with only 0.08% leakage into smaller modes.
Further preliminary experiments on 2D synthetic data and 3D medical image data (hippocampus shapes) are described in [1] and [2], but are omitted here for brevity. Further details and results of this work are published in [1] and [2].
Section 2: Particle-Based Correspondences for Genetic Phenotyping
In cooperation with researchers from the Mario Capecchi lab at the University of Utah, we applied the proposed shape modeling framework from Section 1 to a phenotypic study of several of the forelimb bones of mice with a targeted disruption of the Hoxd11 gene, which Boulet, Davis and Capecchi have previously shown is important for the normal development and patterning of the appendicular skeleton [A. Davis, et al., Development, V. 120, 1995]. Our phenotyping study serves as a further evaluation of the particlebased technology described in the previous section, and is among the first to explore the use of high-dimensional, point-based shape modeling and analysis for small animal phenotyping.
Our study focused on three forelimb bones segmented from the CT volumes of 20 male Hoxd11 -/- (mutant) mice and 20 male wild-type mice: the metacarpal (MC), the first phalange (P1), and the second phalange (P2) of digit 2 of the right forepaw of the mouse. We computed combined models (mutant + wild type) of each of the three bones using the particle-system method described in the previous section for each of the three bones of interest, using 1024 correspondence points per shape. The results from the Hoxd11 study suggest that point-based shape modeling can be an effective tool for the study of mouse skeletal phenotype. We detected global and local group differences between normals and the Hoxd11 mutants that are not observable with traditional metrics, and showed how we can use general linear models to control for covariates in global, univariate shape statistics.
The results of this work are published in [3].
Section 3: Hypothesis Testing and Visualization
In the mouse phenotyping work of the previous section, we described a statistical analysis primarily based on univariate shape measures. In this work we expand our hypothesis testing methodology to incorporate multivariate shape features, and explore methods to aid in the interpretation of statistical results and group differences. Analysis of point-based shape models is difficult because point-wise statistical tests are require multiple-comparisons corrections that significantly reduce statistical power. Analysis in the full shape space, however, is problematic due to the high number of dimensions and the difficulty of obtaining sufficient samples. A common solution is to employ dimensionality reduction reduction by choosing a subspace in which to project the data for traditional multivariate analysis, such as a nonparametric Hotelling T2 test. Dimensionality reduction, however, requires both the choice of the type of basis function to use, and also the basis vector, and the choice of how many basis vectors to use. This latter consideration can be hard to resolve when the choice of different numbers of factors leads to different statistical results.
For our multivariate metric of shape, we propose to use a standard, data-driven approach to dimensionality reduction and project the correspondences into a lower dimensional space determined by choosing a number of basis vectors from principal component analysis (PCA). Ideally, we would like to choose only PCA modes that account for variance that cannot be explained by random noise. Parallel analysis is commonly recommended for this purpose [see, e.g., L W Glorfeld, Educational and Psych. Measurement, V. 55, 1955], which works by comparing the percent variances of each of the PCA modes with the average percent variances obtained via PCA of Monte Carlo simulations of samplings from isotropic, multivariate, unit Gaussian distributions. We choose only modes with greater variance than the simulated modes for the dimensionality reduction, and use use a standard, parametric Hotelling T2 test to test for group differences, with the null hypothesis that the two groups are drawn from the same distribution.
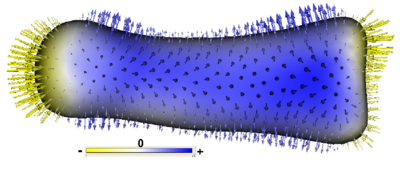
To visualize group differences that are driving the statistical result, we compute the linear discriminant vector implicit in the Hotelling T2 statistic, which is the is the line along which the between-class variance is maximized with respect to the within-class variance (Fisher’s linear discriminant). The discriminant vector can be rotated back from PCA space into the full dimensional shape space, and then mapped onto the mean group shape visualizations to give an indication of the significant morphological differences between groups. Figure 2 shows a linear discriminant visualization on a mean surface of the P2 bone from the Hoxd11 study in the previous section.
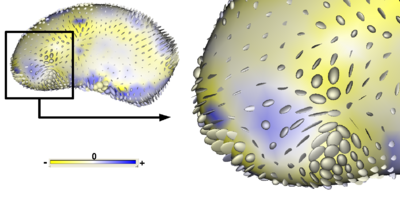
To visualize deformations between the group mean shapes, we can compute metrics on the displacement field describing the mapping from points x on one group mean to corresponding points x0 on the another. Using the set of correspondence points, a smooth transformation T(x) = x0, can be computed using a thin-plate spline interpolation. We propose to visualize strain, a measure on the Jacobian of the deformation field x − T(x) that describes the local stretching and compression caused by the deformation. An effective visualization for the strain tensor is an ellipsoid with principal axes given by the principal directions and magnitude of strain. Figure 3 is a visualization of the strain tensors computed from the deformation from the mean patient shape to the mean normal control shape for putamen shape data.
The results of this work are published in [4].
Section 4: Correspondence Based on Functions of Position
We have extended the particle-system modeling algorithm described in Section 1 to establish correspondence by minimization of the entropy of arbitrary vector-valued functions of position, versus only considering positional information itself. This more general shape modeling algorithm is useful for cases where the notion of correspondence is not well-defined by surface landmarks or topology, but may be better described by other measures. Correspondence computation on populations of human cortical surfaces is one such example because of the high variability of cortical folding patterns across subjects. This work was conducted jointly with Ipek Oguz and Martin Styner from UNC Chapel Hill.
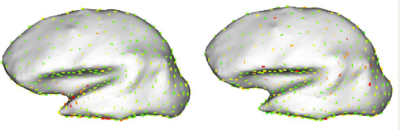
As a test of the above methodology, Oguz, et al. used sulcal depth to optimize correspondence position on 10 cortical surfaces from health patients, reconstructed from T1 images and inflated using FreeSurfer. The study then compared the variance of sulcal depth and cortical thickness measures at optimized correspondence point positions with their variance before optimization, after optimization by the positional method in Section 1, and correspondences chosen by the FreeSurfer algorithm. For sulcal depth measurements, the proposed method reduced variance almost 75-fold over initial data, and almost 25-fold over FreeSurfer results. For cortical thickness, the method showed considerably reduced variance over the unoptimized and positionally-optimized correspondences, with a slightly higher variance than FreeSurfer. An inspection of the distribution of the mean variance over the surface, as shown in Figure 4, reveals that the variance in the FreeSurfer data is more widely distributed across the surface, while for the proposed method, the variance has only a more localized variance around the temporal lobe, which Oguz, et al. note suffered from reconstruction problems due to noise in the input images.
The results of this work are published in [5].
Section 5: Correspondence for Multiple-Object Complexes
We define a multi-object complex as a set of solid shapes, each representing a single, connected biological structure, assembled into a scene within a common coordinate frame. Because the particle-based correspondence method described in Section 1 does not rely on connectivity among correspondences, it can be directly applied to multi-object complexes by treating all of the objects in the complex as one. However, if the object-level correspondence is known a priori, we can assign each particle to a specific object, decouple the spatial interactions between particles on different shapes, and constrain each particle to its associated object, thereby ensuring that each correspondence stays on a particular anatomical structure. The shape-space statistics remain coupled, however, and the covariance matrix in the entropy estimation includes all particle positions across the entire complex, so that optimization takes place on the joint, multi-object model. This work is the first method to demonstrate such a joint optimization of a multi-object shape model.
As an experimental analysis of the proposed methodology, we used multi-object segmentation data taken from an ongoing longitudinal pediatric autism study [Hazlett, et al. Arch Gen Psych, V. 62, 2005]. The data consists of 10 subcortical brain structures derived from MRI brain scans of autistic subjects (10 samples) and typically-developing controls (15 samples) at 2 years of age (see Fig 6). We sampled each complex of segmentations with 10,240 correspondence points, using 1024 particles per structure. For comparison, we also computed point-correspondence models for each of the 10 structures separately and concatenated their correspondences together to form a marginally optimized joint model. We found significant differences in structure scale only for the right and left amygdala, and no significant group differences in structure position or pose. The hypothesis test method outlined in Section 3 gives a highly significant p-value of 0.0087, which is the first evidence shown for this data for group differences in shape alone (i.e. without scale). By contrast hypothesis testing with the marginally-optimized model showed a significant decrease in statistical power, with a p-value of 0.0480. Figure 6 shows the mean shape surfaces for the normal and autistic groups, as reconstructed from the Euclidean averages of the correspondence points. Each structure is displayed in its mean orientation, position, and scale in the global coordinate frame. We computed the average orientation for each structure using methods for averaging in curved spaces. The length in the surface normal direction of each of the point-wise discriminant vector components for the autism data is given by the colormap.
The results of this work are published in [6].
Key Investigators
Utah: Joshua Cates, Tom Fletcher, Ross Whitaker
Publications
- J Cates, P T Fletcher, Z Warnock, and R Whitaker, A shape analysis framework for small animal phenotyping with application to mice with a targeted disruption of Hoxd11, in Proc. 5th IEEE International Symposium on Biomedical Imaging (ISBI ’08), 2008, pp. 512–516.
- J Cates, PT Fletcher, and R Whitaker, “A hypothesis testing framework for high-dimensional shape models,” in Mathematical Foundations of Computational Anatomy Workshop. 2008, pp. 170–181, MICCAI.
- I Oguz, J Cates, P T Fletcher, Z Warnock, R Whitaker, D Cool, S Aylward, and M Styner, “Cortical correspondence using entropy-based particle systems and local featrues,” in Proc. 5th IEEE International Symposium on Biomedical Imaging (ISBI ’08), 2008, pp. 1637–1640.
- J Cates, PT Fletcher, M Styner, H Hazlett, and R Whitaker, “Particle-based shape analysis of multi-object complexes,” in Proceedings of the 11th International Conference on Medical Image Computing and Computer Assisted Intervention. (MICCAI 08).