Projects:QIN:3D Slicer Annotation Image Markup:Standard Terminology
This is a collection of relevant pieces from:
Contents
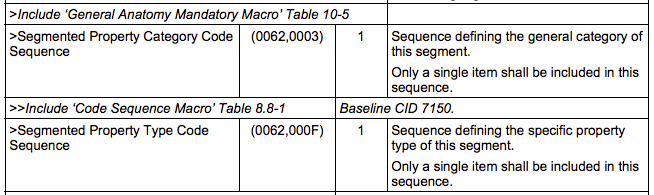
Segmentation image module attributes
Segmentation image module attributes must in particular include the following (PS 3.3-2011, p.989):
Code sequence attributes
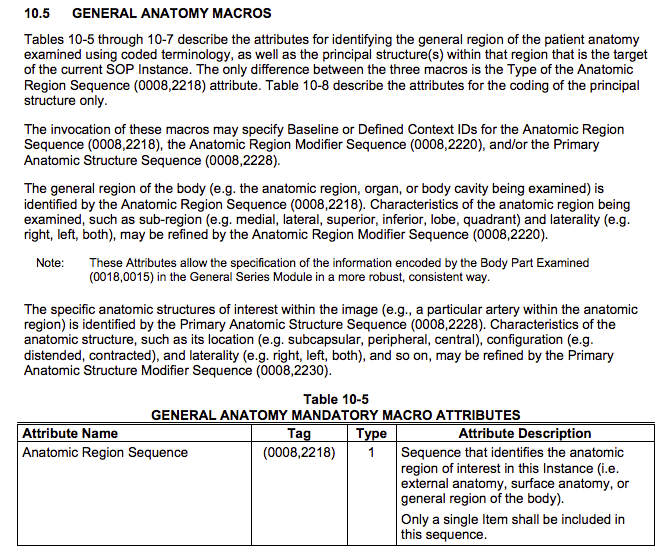
General Anatomy Macro
The General Anatomy Macro is defined as follows (PS 3.3-2011, p.96):
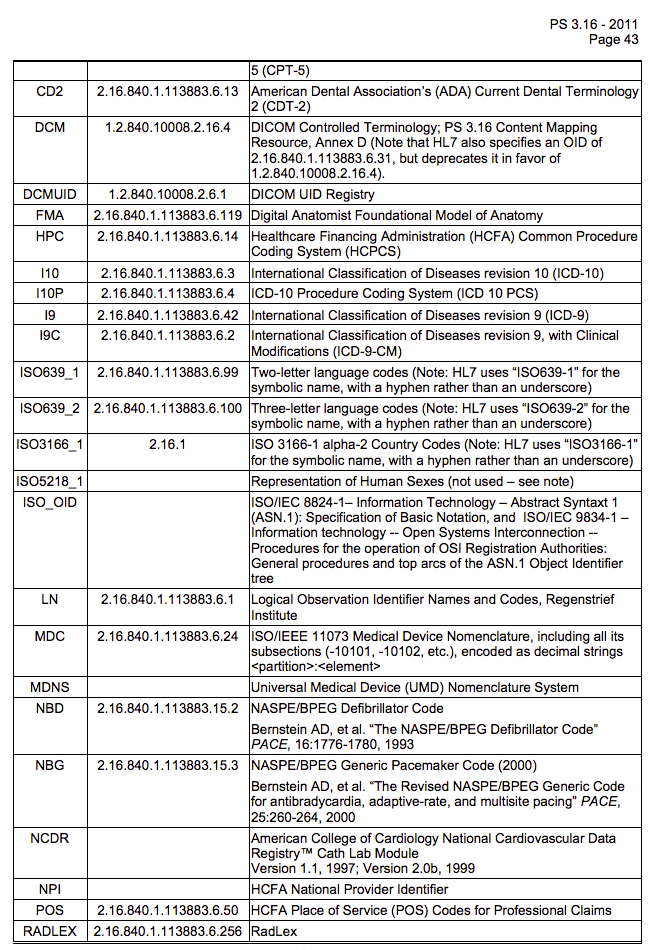
Coding Scheme Designators
NOTE: this table is incomplete, consult PS 3.16-2011 p.43.
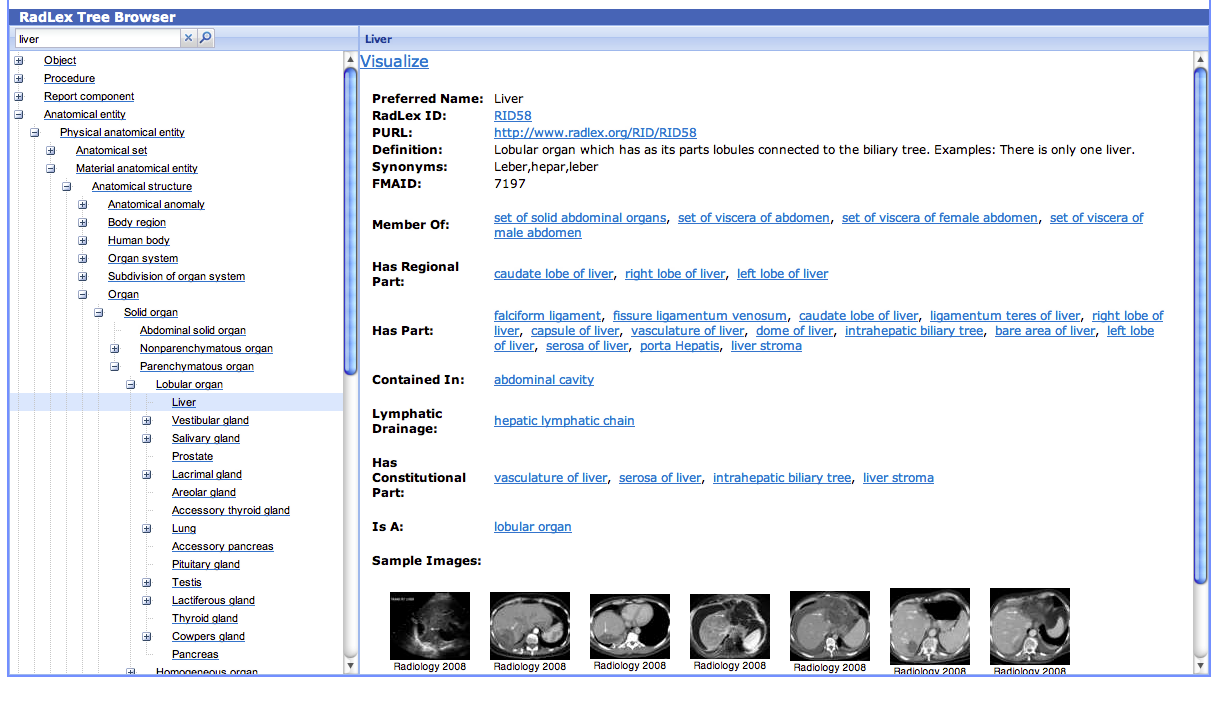
RadLex
- General information: http://rsna.org/RadLex.aspx
- License: http://rsna.org/uploadedFiles/RSNA/Content/Informatics/radlex_public_license_version_1-0-1.pdf
- Example hierarchy of the term mapping from RadLex browser (liver: http://www.radlex.org/RID/RID13419):
SNOMED
David Clunie in email communication Sept 17, 2012:
[...] The [DICOM] agreement with SNOMED is to allow any implementer to use the terms included in DICOM in their implementations and products without fee; it is not just to be able to include them in the text of the standard documents. So, since Slicer would be using the concepts to include in a DICOM SEG object, you do not need any additional agreement, nor any agreement with SNOMED to distribute the source code (my PixelMed Java DICOM toolkit is full of SNOMED codes, for example, and I neither have not need such an additional agreement).
Notes on choosing SNOMED concepts when fleshing out the table, esp. regarding anatomy. Make sure that you choose "(body structure)" concepts for anatomy, and that you pick the "entire ..." concept rather than "structure of" etc., and if the choice is not obvious, include about your choice so that these can be flagged for special attention during review.
One way to find the right SNOMED code without using one of the terminology browsers is just to download the text files from the UMLS (from http://www.nlm.nih.gov/research/umls/licensedcontent/snomedctfiles.html). You will need to register and get a user name and password.
Then you can use grep as follows:
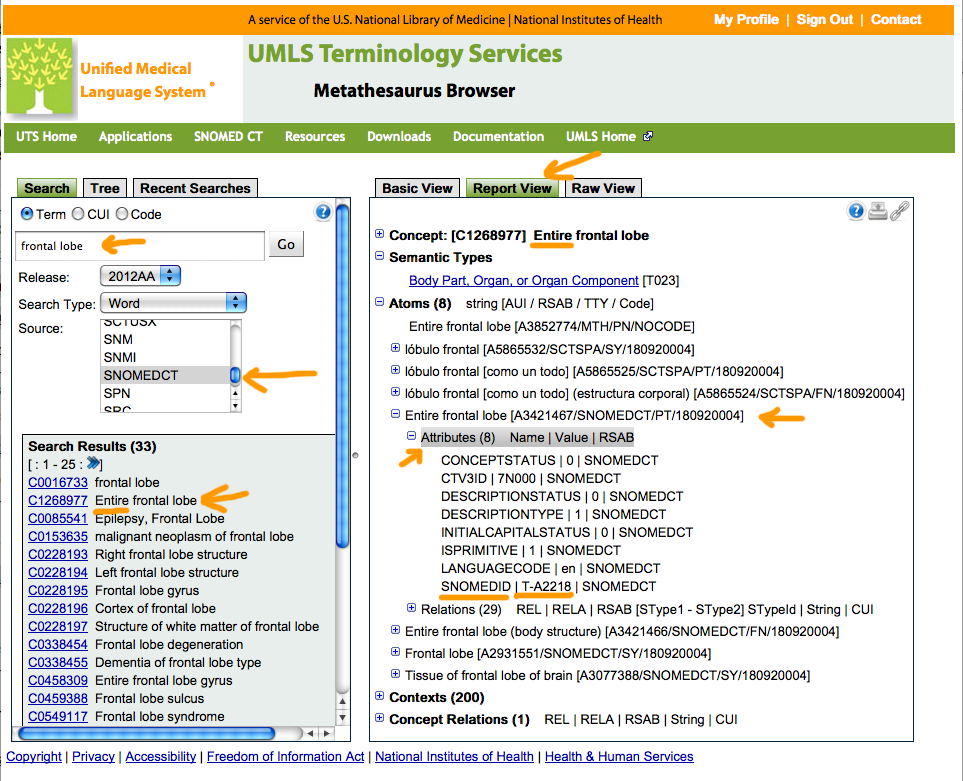
% grep -i 'frontal lobe' sct1_Concepts_Core_INT_20110131.txt | grep -i structure | grep -i entire 180920004 0 Entire frontal lobe (body structure) 7N000 T-A2218 1 279169003 0 Entire frontal lobe gyrus (body structure) Xa10B T-A2207 1 314141005 0 Entire right frontal lobe (body structure) XaEYz T-A2213 0 314142003 0 Entire left frontal lobe (body structure) XaEZ0 T-A2214 0 362330001 0 Entire cortex of frontal lobe (body structure) XU96w T-A2211 1 362331002 0 Entire white matter of frontal lobe (body structure) XU96y T-A2221 1
You can find this sort of thing using the UMLS Terminology Browser (at https://uts.nlm.nih.gov/metathesaurus.html), for example, as follows:
Mapping Slicer GeneralAnatomy LUT to Standard Terminology (work in progress)
Original GeneralAnatomy LUT see here: http://www.slicer.org/slicerWiki/index.php/Documentation/4.1/SlicerApplication/LookupTables/GenericAnatomyColors
Standing issues: licensing and redistribution
TODO items: consider using alternative terminology vocabularies (will add corresponding columns):
- FMA
- browser http://fme.biostr.washington.edu/FME/
- available under CreativeCommons license: http://sig.biostr.washington.edu/projects/fm/index.html http://sig.biostr.washington.edu/projects/fma/release/index.html
- Medical Subject Headings (MeSH)
- NeuroLex: http://neurolex.org/wiki/Main_Page (Karl Helmer from MGH used it and can help with the translation effort)
NOTE: work in progress translating Slicer General Anatomy into SNOMED terminology is done on a Google Documents spreadsheet. Please email Andrey Fedorov (fedorov at Bwh (.) harvard edu) if you are interested to participate or see the status.
Segmentation objects allow to encode recommended display color in "Recommended Display CIELab Value" (0062,000D) attribute; these values are encoded in CIALab.