Difference between revisions of "Projects:RegistrationDocumentation:ParameterTesting"
From NAMIC Wiki
| (33 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | === | + | === Concept / Design === |
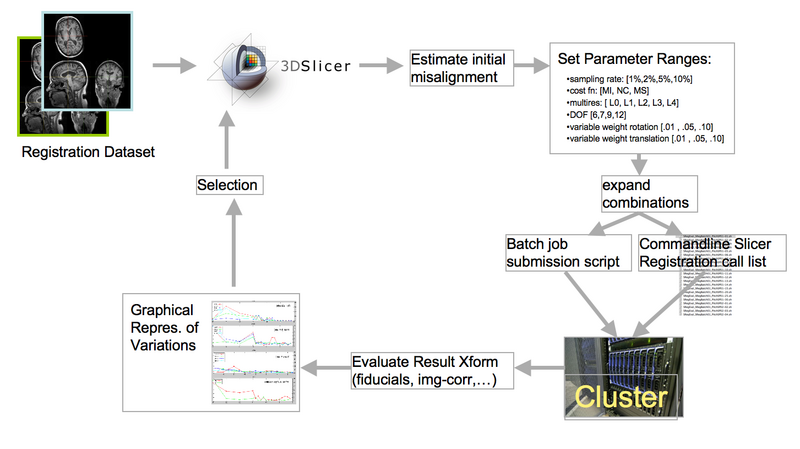
| + | [[Image:RegLib_ParameterExploration_Schematic.png|800px|lleft|basic schematic of searching for the best possible registration via batch submission of parameter exploration]] | ||
| + | |||
| + | === Commandline Setup === | ||
| + | *You can run the RegisterImages module from the command-line. For downloaded pre-compiled versions of 3Dslicer, the the LD library (or DYLD for Mac) paths must be set. For example, add to your .bashrc file | ||
| + | # enable slicer commandline for RegisterImages | ||
| + | export DYLD_LIBRARY_PATH=$DYLD_LIBRARY_PATH:/Applications/Slicer3-3.5a/lib/InsightToolkit:/Applications/Slicer | ||
| + | 3-3.5a/lib/MRML/:/Applications/Slicer3-3.5a/lib/vtkITK:/Applications/Slicer3-3.5a/lib/vtk-5.4:/Applications/Sl | ||
| + | icer3-3.5a/lib/FreeSurfer:/Applications/Slicer3-3.5a/lib/libvtkTeem.dylib:/Applications/Slicer3-3.5a/lib/vtkTe | ||
| + | em:/Applications/Slicer3-3.5a/lib/Teem-1.10.0 | ||
| + | *a commandline call then looks like this: | ||
| + | YourSlicerHome/lib/Slicer3/Plugins/RegisterImages FixedImageName MovingImageName --registration PipelineRigid | ||
| + | --initialization ImageCenters --metric MattesMI --rigidSamplingRatio 0.02 --saveTransform OutputXformName.tfm | ||
| + | *to get all the commandline options type | ||
| + | YourSlicerHome/lib/Slicer3/Plugins/RegisterImages -h | ||
| + | |||
| + | === Commandline Compilation of New Kitware Multiresolution Code === | ||
| + | *download: CMake, svn, Xcode, | ||
| + | *within dedicated source directory, call | ||
| + | svn update | ||
| + | *within dedicated binary directory, call | ||
| + | make | ||
| + | *call | ||
| + | ./MoreSearch S027_SPGR15.nii S027_SPGR15_aff0.nii ResOut.nii | ||
| + | * currently there is no input or output Xform. | ||
| + | * action item: align | ||
| + | * discussion 11-20: will add input & output Xform for evaluation. | ||
| + | |||
| + | === Parameter Exploration on Cluster SOP === | ||
| + | * Matlab script ''SRegEval_BatchBuild'' generates input transforms, individual and master submission shell scripts and directory structures for batch submission on linux cluster | ||
| + | * tar and Xfer to cluster | ||
| + | * untar and run submission script. Each test registration becomes 1 submission. | ||
| + | * tar result structure and Xfer back to project directory | ||
| + | * Matlab script ''RegEval'' evaluates result Xfoms within ICC and builds RMS plots and rank statistics | ||
| + | * to be explored: use of GWE for batch submission and review | ||
| + | * use of BatchMake for batch preparation | ||
| + | === Pending: Parameter Exploration via GWE === | ||
| + | *[http://www.gridwizard.org/ GridWizard Main Site] | ||
| + | |||
| + | === Pending: Parameter Exploration via BatchMake === | ||
| + | *[http://www.batchmake.org/Wiki/BatchMake BatchMake intro] | ||
| + | *[http://www.batchmake.org/ Main BatchMake site] | ||
| + | *[http://www.batchmake.org/Wiki/Batchmake_tutorial#Running_an_application_with_BatchMake use BatchMake as a stand-alone application] | ||
| + | *[http://www.batchmake.org/Wiki/Batchmake_slicer use BatchMake inside Slicer] | ||
| + | |||
| + | === Parameter Exploration Abstract 2010 === | ||
*'''Title:''' Protocol-Tailored Automated MR Image Registration | *'''Title:''' Protocol-Tailored Automated MR Image Registration | ||
| − | *'''Authors:''' Dominik Meier, Casey Goodlett, Ron Kikinis | + | *'''Authors:''' Dominik Meier, Casey Goodlett, Ron Kikinis |
| − | *'''Significance:''' Fast and accurate algorithms for automated co-registration of different MR datasets are commonly available. But systematic knowledge or guidelines for choosing registration strategies and parameters for these algorithms | + | *'''Significance:''' Fast and accurate algorithms for automated co-registration of different MR datasets are commonly available. But systematic knowledge or guidelines for choosing registration strategies and parameters for these algorithms are lacking. As image contrast and initial misalignment vary, so do the appropriate choices of cost function, initialization and optimization strategy. Beyond using the defaultt settings, the research end-user is forced to resort to trial and error to find a successful registration, which is ineffcient and yields suboptimal results. Registration problems also contain an inherent tradeoff between accuracy and robustness, and the landscape of this tradeoff also is largely unknown. We present guidelines for the end-user to choose parameters for registration of MR images of different contrasts and resolution. This is part of a concerted effort to build a Registration Case Library available to the medical imaging research community. This first batch determines guidelines for affine registration of intra-subject multi-contrast brain MRI data within the 3DSlicer software. |
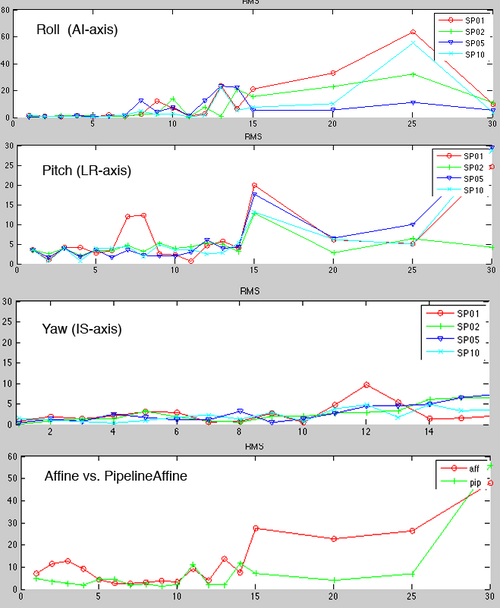
| − | + | [[Image:SRegVar_Roll1.png|right|500px|360 Level Self Validation Test: FLAIR to T1, 1,2,5,10% sampling rates]] | |
| + | [[Image:SRegTest_FLAIR-T1_LRRot15.png|right|500px|360 Level Self Validation Test: FLAIR to T1, left-right rotation]] | ||
*'''Method:''' | *'''Method:''' | ||
| − | ** | + | **xx brain MRI pairs from 4 different subjects with optimal registrations were selected. The pairs represent all 6 contrast combinations of T1,T2,FLAIR and PD. Each pair was then disturbed by a known validation transform and subsequently registered again under different parameter settings. Registration success was evaluated as the extent to which the initial position could be recovered, reported as distributions of RMS of all points within the brain. Included in the variations were 1) degree of initial misalignment (18 levels), 2) cost function (MI, NCorr, MSq), 3) %-sampling ratio (1%,2%,5%), 4) masking , 5) low-DOF prealignment, 6) initialization/start position (none, image center, center of mass, 2nd moment). The parameter set that performed best across the range of misalignment is documented. We choose 3-4 subjects/exams with 3-4 different contrast pairings: T1, T2, PD, FLAIR: ~12-16 images. We choose sets for which we have a good registration solution. Each validation test comprised the following: |
| − | + | **1- disturb each pair by a known transform of varying rotational & translational misalignment. | |
| − | + | **2- run registration for a set of parameter settings and save the result Xform, e.g. metric: NormCorr vs. MI | |
| − | ** | + | **3- evaluate registration error as RMS of point distance for all points within the brain. |
| − | ** | + | **4- run sensitivity analysis and report the best performing parameter set for each MR-MR combination |
| − | ** | ||
| − | ** | ||
| − | |||
*This self-validation scheme avoids recruiting an expert reader to determine ~ 3-5 anatomical landmarks on each unregistered image pair (time constraint). Also we can cover a wider range of misalignment and sensitivity by controlling the input Xform. It also facilitates batch processing. | *This self-validation scheme avoids recruiting an expert reader to determine ~ 3-5 anatomical landmarks on each unregistered image pair (time constraint). Also we can cover a wider range of misalignment and sensitivity by controlling the input Xform. It also facilitates batch processing. | ||
*'''Results:''' | *'''Results:''' | ||
| − | ** | + | **Performance is relative insensitive to parameter permutations if misalignment is below 5 degrees, i.e. for small adjustments many settings yield success |
| − | ** | + | **Above 5 degrees parameter influence becomes non-contiguous: as shown by results small variation in input parameter can produce drastically different results. |
| − | ** | + | **Registration failure sets in, disregarding of (singular) parameter settings, above 15 degrees rotational misalignment. |
| − | [[Image: | + | **most sensitive is Pitch (LR-axis rotation) or head tilt. Small rotations here are significantly more likely to cause misalignment than other angles. |
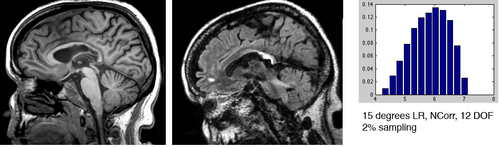
| − | [[Image:RegistrationISMRM10_Disturb_12_hist.png|left|300px|RMS Histogram comparison for 12 degrees misalignment]] | + | **higher sampling rates do not necessarily promise better results, but 2% seems a lower bound unless speed is a critical issue |
| − | [[Image:RegistrationISMRM10_RMS.png|left|300px|Sensitivity (RMS range) vs. rotational misalignment]] | + | **PipelineAffine outperforms Affine in almost all instances. |
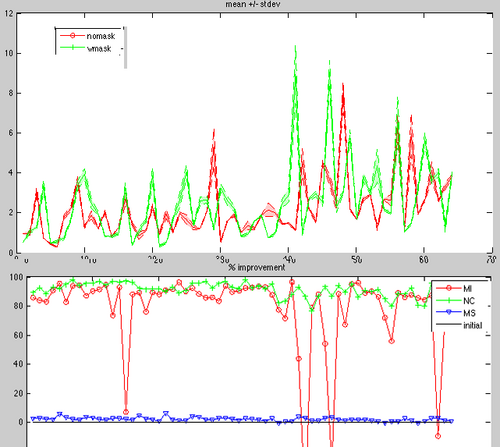
| + | **UPDATE: Metric comparison: mutual Info, norm corr, msqrd | ||
| + | **UPDATE: masking comparison: effect of masking on robustness | ||
| + | **UPDATE: initialization (none, image center, center of mass, 2nd moment) | ||
| + | **UPDATE: search step sizes (rotational, translational) : have an effect but could be considered heuristics that ought to be determined by the algorithm | ||
| + | **UPDATE: Bias field comparison: at what level of inhomogeneity does performance degrade. Which cost function is more robust against inhomogeneity? | ||
| + | *'''Discussion:''' | ||
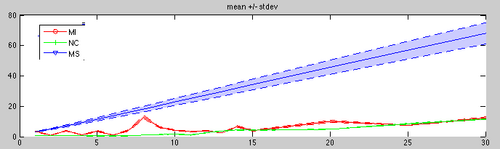
| + | **Metric comparison: (status Nov.4): NormCorr not parallelized and therfore slow, but best quality. MSqrd does not move. | ||
| + | [[Image:sRegVar_MSqrdError.png|left|500px|Metric comparison]] | ||
| + | [[Image:SRegEval_CostFn+Masking.png|right|500px|Metric comparison]] | ||
| + | | ||
| + | <!-- [[Image:RegistrationISMRM10_Disturb_12_hist.png|left|300px|RMS Histogram comparison for 12 degrees misalignment]] --> | ||
| + | <!-- [[Image:RegistrationISMRM10_RMS.png|left|300px|Sensitivity (RMS range) vs. rotational misalignment]]--> | ||
Latest revision as of 15:52, 23 November 2009
Home < Projects:RegistrationDocumentation:ParameterTestingBack to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
Concept / Design
Commandline Setup
- You can run the RegisterImages module from the command-line. For downloaded pre-compiled versions of 3Dslicer, the the LD library (or DYLD for Mac) paths must be set. For example, add to your .bashrc file
# enable slicer commandline for RegisterImages export DYLD_LIBRARY_PATH=$DYLD_LIBRARY_PATH:/Applications/Slicer3-3.5a/lib/InsightToolkit:/Applications/Slicer 3-3.5a/lib/MRML/:/Applications/Slicer3-3.5a/lib/vtkITK:/Applications/Slicer3-3.5a/lib/vtk-5.4:/Applications/Sl icer3-3.5a/lib/FreeSurfer:/Applications/Slicer3-3.5a/lib/libvtkTeem.dylib:/Applications/Slicer3-3.5a/lib/vtkTe em:/Applications/Slicer3-3.5a/lib/Teem-1.10.0
- a commandline call then looks like this:
YourSlicerHome/lib/Slicer3/Plugins/RegisterImages FixedImageName MovingImageName --registration PipelineRigid --initialization ImageCenters --metric MattesMI --rigidSamplingRatio 0.02 --saveTransform OutputXformName.tfm
- to get all the commandline options type
YourSlicerHome/lib/Slicer3/Plugins/RegisterImages -h
Commandline Compilation of New Kitware Multiresolution Code
- download: CMake, svn, Xcode,
- within dedicated source directory, call
svn update
- within dedicated binary directory, call
make
- call
./MoreSearch S027_SPGR15.nii S027_SPGR15_aff0.nii ResOut.nii
- currently there is no input or output Xform.
- action item: align
- discussion 11-20: will add input & output Xform for evaluation.
Parameter Exploration on Cluster SOP
- Matlab script SRegEval_BatchBuild generates input transforms, individual and master submission shell scripts and directory structures for batch submission on linux cluster
- tar and Xfer to cluster
- untar and run submission script. Each test registration becomes 1 submission.
- tar result structure and Xfer back to project directory
- Matlab script RegEval evaluates result Xfoms within ICC and builds RMS plots and rank statistics
- to be explored: use of GWE for batch submission and review
- use of BatchMake for batch preparation
Pending: Parameter Exploration via GWE
Pending: Parameter Exploration via BatchMake
- BatchMake intro
- Main BatchMake site
- use BatchMake as a stand-alone application
- use BatchMake inside Slicer
Parameter Exploration Abstract 2010
- Title: Protocol-Tailored Automated MR Image Registration
- Authors: Dominik Meier, Casey Goodlett, Ron Kikinis
- Significance: Fast and accurate algorithms for automated co-registration of different MR datasets are commonly available. But systematic knowledge or guidelines for choosing registration strategies and parameters for these algorithms are lacking. As image contrast and initial misalignment vary, so do the appropriate choices of cost function, initialization and optimization strategy. Beyond using the defaultt settings, the research end-user is forced to resort to trial and error to find a successful registration, which is ineffcient and yields suboptimal results. Registration problems also contain an inherent tradeoff between accuracy and robustness, and the landscape of this tradeoff also is largely unknown. We present guidelines for the end-user to choose parameters for registration of MR images of different contrasts and resolution. This is part of a concerted effort to build a Registration Case Library available to the medical imaging research community. This first batch determines guidelines for affine registration of intra-subject multi-contrast brain MRI data within the 3DSlicer software.
- Method:
- xx brain MRI pairs from 4 different subjects with optimal registrations were selected. The pairs represent all 6 contrast combinations of T1,T2,FLAIR and PD. Each pair was then disturbed by a known validation transform and subsequently registered again under different parameter settings. Registration success was evaluated as the extent to which the initial position could be recovered, reported as distributions of RMS of all points within the brain. Included in the variations were 1) degree of initial misalignment (18 levels), 2) cost function (MI, NCorr, MSq), 3) %-sampling ratio (1%,2%,5%), 4) masking , 5) low-DOF prealignment, 6) initialization/start position (none, image center, center of mass, 2nd moment). The parameter set that performed best across the range of misalignment is documented. We choose 3-4 subjects/exams with 3-4 different contrast pairings: T1, T2, PD, FLAIR: ~12-16 images. We choose sets for which we have a good registration solution. Each validation test comprised the following:
- 1- disturb each pair by a known transform of varying rotational & translational misalignment.
- 2- run registration for a set of parameter settings and save the result Xform, e.g. metric: NormCorr vs. MI
- 3- evaluate registration error as RMS of point distance for all points within the brain.
- 4- run sensitivity analysis and report the best performing parameter set for each MR-MR combination
- This self-validation scheme avoids recruiting an expert reader to determine ~ 3-5 anatomical landmarks on each unregistered image pair (time constraint). Also we can cover a wider range of misalignment and sensitivity by controlling the input Xform. It also facilitates batch processing.
- Results:
- Performance is relative insensitive to parameter permutations if misalignment is below 5 degrees, i.e. for small adjustments many settings yield success
- Above 5 degrees parameter influence becomes non-contiguous: as shown by results small variation in input parameter can produce drastically different results.
- Registration failure sets in, disregarding of (singular) parameter settings, above 15 degrees rotational misalignment.
- most sensitive is Pitch (LR-axis rotation) or head tilt. Small rotations here are significantly more likely to cause misalignment than other angles.
- higher sampling rates do not necessarily promise better results, but 2% seems a lower bound unless speed is a critical issue
- PipelineAffine outperforms Affine in almost all instances.
- UPDATE: Metric comparison: mutual Info, norm corr, msqrd
- UPDATE: masking comparison: effect of masking on robustness
- UPDATE: initialization (none, image center, center of mass, 2nd moment)
- UPDATE: search step sizes (rotational, translational) : have an effect but could be considered heuristics that ought to be determined by the algorithm
- UPDATE: Bias field comparison: at what level of inhomogeneity does performance degrade. Which cost function is more robust against inhomogeneity?
- Discussion:
- Metric comparison: (status Nov.4): NormCorr not parallelized and therfore slow, but best quality. MSqrd does not move.