Projects:RegistrationDocumentation:ParameterTesting
From NAMIC Wiki
Home < Projects:RegistrationDocumentation:ParameterTesting
Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
ISMRM abstract 2010
- Title: Protocol-Tailored Automated MR Image Registration
- Authors: Dominik Meier, Casey Goodlett, Ron Kikinis
- Significance: Fast and accurate algorithms for automated co-registration of different MR datasets are commonly available. But systematic knowledge or guidelines for choosing registration strategies and parameters for these algorithms is lacking. As image contrast and initial misalignment vary, so do the appropriate choices of cost function, initialization and optimization strategy. To find a successful registration the clinical research end user is currently forced to resort trial and error, which is ineffcient and yields suboptimal results. Registration problems also contain an inherent tradeoff between accuracy and robustness, and the landscape of this tradeoff also is largely unknown.
- Objective: Determine guidelines for affine registration of intra-subject multi-contrast MRI data within the 3DSlicer software. This work will present approaches and solutions for successful registration for a large set of combinations of MRI pairings. This is part of a concerted effort to build a Registration Case Library available to the medical imaging research community.
- Method:
- 1- choose 3-4 subjects/exams with 3-4 different contrast pairings: T1, T2, PD, FLAIR: ~12-16 images. We choose sets for which we have a good registration solution
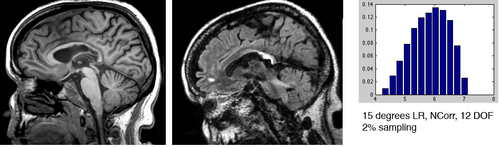
- 2- disturb each pair by a known transform of varying rotational & translational misalignment
- 3- run registration for a set of parameter settings and save the result Xform, e.g. metric: NormCorr vs. MI , 2% vs 5% sampling, 50 vs. 100 iteration max
- 4- evaluate registration error as point distance and RMS.
- 5- run sensitivity analysis and report the best performing parameter set for each MR-MR combination
- 6-extension 1: add different voxel sizes, i.e. emulate 1,3,5mm slice thickness
- This self-validation scheme avoids recruiting an expert reader to determine ~ 3-5 anatomical landmarks on each unregistered image pair (time constraint). Also we can cover a wider range of misalignment and sensitivity by controlling the input Xform. It also facilitates batch processing.
- Results:
- range of fiducial misalignment & distributions
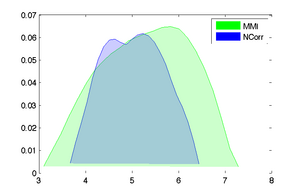
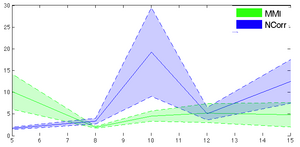
- Plot error vs. initial misalignment (where does registration begin to fail), plot error vs. parameter settings (which setting works best for the toughest case)
- Examples below are cost function comparisons: MI vs NCorr