Difference between revisions of "Projects:RegistrationEvaluation"
Jfishbaugh (talk | contribs) |
Jfishbaugh (talk | contribs) |
||
| Line 6: | Line 6: | ||
can be determined. | can be determined. | ||
| − | = Example 1: Fixing Artifacts in DWI = | + | = Example 1: Fixing Artifacts in DWI with Slice by Slice 2D Registration = |
The DWI data consists of 13 volumes -- a b0 and 12 gradient directions. This process was conducted 3 times per patient for a total of 39 volumes. The image below shows a particular | The DWI data consists of 13 volumes -- a b0 and 12 gradient directions. This process was conducted 3 times per patient for a total of 39 volumes. The image below shows a particular | ||
| Line 20: | Line 20: | ||
Using ITK modules, we wrote an application to perform 2D affine transformation with the following parameters: | Using ITK modules, we wrote an application to perform 2D affine transformation with the following parameters: | ||
| − | :'''Similarity Metric''': | + | :'''Similarity Metric''': MattesMutualInformationImageToImageMetric with 24 bins and 10000 samples per bin. |
| − | :'''Interpolation''': | + | :'''Interpolation''': LinearInterpolateImageFunction |
| − | :'''Optimization''': | + | :'''Optimization''': RegularStepGradientDescentOptimizer |
[[Image:itk_reg.png|300px]] | [[Image:itk_reg.png|300px]] | ||
Revision as of 00:40, 7 October 2009
Home < Projects:RegistrationEvaluationContents
Evaluation of Registration
We are interested in comparing existing registration packages to determine how registration in Slicer3 can be improved. This work focuses on examining various packages researchers are currently using for registration and comparing results on a set of examples representative of common registration tasks. Finally, we propose the development of a testbed, where registration packages can be compared and parameters suitable for a given domain (e.g. multi-modal brain registration) can be determined.
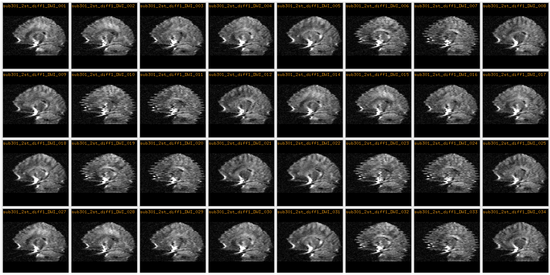
Example 1: Fixing Artifacts in DWI with Slice by Slice 2D Registration
The DWI data consists of 13 volumes -- a b0 and 12 gradient directions. This process was conducted 3 times per patient for a total of 39 volumes. The image below shows a particular slice in all 12 gradient directions and all 3 scans where the artifacts can clearly be seen in some of the gradient directions.
The problem can be alleviated by 2D affine registration between slices of a corrupted volume with the corresponding slice from the b0 volume, in which we assume the artifact is not present. We now consider different registration packages applied to this problem.
ITK
Using ITK modules, we wrote an application to perform 2D affine transformation with the following parameters:
- Similarity Metric: MattesMutualInformationImageToImageMetric with 24 bins and 10000 samples per bin.
- Interpolation: LinearInterpolateImageFunction
- Optimization: RegularStepGradientDescentOptimizer