Difference between revisions of "Projects:RegistrationLibrary:RegLib C02"
From NAMIC Wiki
m (→Keywords) |
|||
| Line 19: | Line 19: | ||
This scenario occurs in many forms whenever we wish to align all the series from a single MRI exam/session into a common space. Alignment is necessary because the subject likely has moved in between series. | This scenario occurs in many forms whenever we wish to align all the series from a single MRI exam/session into a common space. Alignment is necessary because the subject likely has moved in between series. | ||
=== Keywords === | === Keywords === | ||
| − | MRI, brain, head, intra-subject, FLAIR, T1, defacing, masking | + | MRI, brain, head, intra-subject, FLAIR, T1, defacing, masking, labelmap, segmentation |
| + | |||
===Input Data=== | ===Input Data=== | ||
*reference: T1 SPGR , 1x1x1 mm voxel size, sagittal, RAS orientation | *reference: T1 SPGR , 1x1x1 mm voxel size, sagittal, RAS orientation | ||
Revision as of 17:20, 20 October 2009
Home < Projects:RegistrationLibrary:RegLib C02Contents
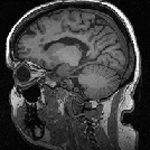
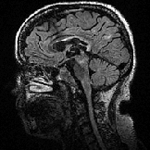
Slicer Registration Use Case Exampe: Intra-subject Brain MR FLAIR to MR T1
|
|

| |
| 1mm isotropic | 1.2mm isotropic | 1.2mm isotropic |
Objective / Background
This scenario occurs in many forms whenever we wish to align all the series from a single MRI exam/session into a common space. Alignment is necessary because the subject likely has moved in between series.
Keywords
MRI, brain, head, intra-subject, FLAIR, T1, defacing, masking, labelmap, segmentation
Input Data
- reference: T1 SPGR , 1x1x1 mm voxel size, sagittal, RAS orientation
- moving: T2 FLAIR 1.2x1.2x1.2 mm voxel size, sagittal, RAS orientation
- quick view JPEG of a lighbox. Does your data look like this?
- download dataset to load into slicer
Registration Challenges
- we expect the amount of misalignment to be small
- we know the underlying structure/anatomy did not change, hence whatever residual misalignment remains is of technical origin.
- the different series may have different FOV. The additional image data may distract the algorithm and require masking
- the different series may have very different resolution and anisotropic voxel sizes
- hi-resolution datasets may have defacing applied to one or both sets, and the defacing-masks may not be available
- the different series may have different contrast.
- individual series may contain motion or other artifacts
Procedure
- step-by step text instruction
- recommended parameter settings
- guided video tutorial
- power point tutorial
Registration Results
- registration parameter presets file (load into slicer and run the registration)
- result transform file (load into slicer and apply to the target volume)
- result screenshots (compare with your results)
- result evaluations (metrics)