Difference between revisions of "Projects:RegistrationLibrary:RegLib C09"
From NAMIC Wiki
(Created page with 'Back to ARRA main page <br> Back to Registration main page <br> [[Projects:RegistrationDocumentation:UseCaseInv…') |
|||
| Line 4: | Line 4: | ||
==Slicer Registration Library Exampe #9: Functional MRI aligned with structural reference MRI== | ==Slicer Registration Library Exampe #9: Functional MRI aligned with structural reference MRI== | ||
| + | [[Image:RegLib C09 fMRI1.png|70px|lleft|RegLib 09: T1 SPGR]] [[Image:RegLib C09 fMRI2.png|70px|lleft|RegLib 09: fMR]] | ||
{| style="color:#bbbbbb; background-color:#333333;" cellpadding="10" cellspacing="0" border="0" | {| style="color:#bbbbbb; background-color:#333333;" cellpadding="10" cellspacing="0" border="0" | ||
| − | |[[Image: | + | |[[Image:RegLib C09 fMRI1.png|150px|lleft|this is the fixed reference image. All images are aligned into this space]] |
|[[Image:Arrow_left_gray.jpg|100px|lleft]] | |[[Image:Arrow_left_gray.jpg|100px|lleft]] | ||
| − | |[[Image: | + | |[[Image:RegLib C09 fMRI2.png|150px|lleft|this is the moving image. The transform is calculated by matching this to the reference image]] |
| − | |||
| − | |||
[[Image:Button_red_fixed.jpg|20px|lleft]] this indicates the reference image that is fixed and does not move. All other images are aligned into this space and resolution<br> | [[Image:Button_red_fixed.jpg|20px|lleft]] this indicates the reference image that is fixed and does not move. All other images are aligned into this space and resolution<br> | ||
[[Image:Button_green_moving.jpg|20px|lleft]] this indicates the moving image that determines the registration transform. <br> | [[Image:Button_green_moving.jpg|20px|lleft]] this indicates the moving image that determines the registration transform. <br> | ||
| − | |||
</small></small> | </small></small> | ||
|- | |- | ||
| − | |[[Image:Button_red_fixed.jpg|40px|lleft]] | + | |[[Image:Button_red_fixed.jpg|40px|lleft]] T1 structural reference |
| | | | ||
| − | |[[Image:Button_green_moving.jpg|40px|lleft]] | + | |[[Image:Button_green_moving.jpg|40px|lleft]] fMRI 4D volume |
| − | |||
|- | |- | ||
|0.46 x 0.46 x 3.0 mm axial <br> 512 x 512 x 46<br>RAS | |0.46 x 0.46 x 3.0 mm axial <br> 512 x 512 x 46<br>RAS | ||
| | | | ||
|1.0 x 1.0 x 3.3 mm <br> axial oblique<br> 256 x 256 x 36<br>RAS | |1.0 x 1.0 x 3.3 mm <br> axial oblique<br> 256 x 256 x 36<br>RAS | ||
| − | |||
|} | |} | ||
===Objective / Background === | ===Objective / Background === | ||
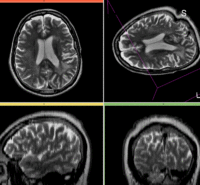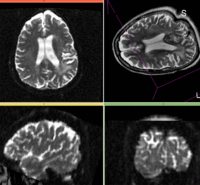
| − | This is a typical example of | + | This is a typical example of fMRI pre-processing. Goal is to align the fMRI image with a structural scan that provides accuracte anatomical reference. The fMRI contains acquisition-related distortion and low contrast to discern much anatomical detail. We also have pathology (stroke) with variable contrast across different MRI protocols. |
=== Keywords === | === Keywords === | ||
| − | MRI, brain, head, intra-subject, | + | MRI, brain, head, intra-subject, fMRI |
===Input Data=== | ===Input Data=== | ||
| − | *[[Image:Button_red_fixed_white.jpg|20px]]reference/fixed : | + | *[[Image:Button_red_fixed_white.jpg|20px]]reference/fixed : T1 |
| − | *[[Image:Button_green_moving_white.jpg|20px]] moving: | + | *[[Image:Button_green_moving_white.jpg|20px]] moving: fMRI sequence of motor task (right hand clench) |
| − | |||
=== Registration Results=== | === Registration Results=== | ||
| Line 64: | Line 59: | ||
=== Discussion: Registration Challenges === | === Discussion: Registration Challenges === | ||
| − | * | + | *the fMRI contains acquisition-related distortions that can make automated registration difficult. |
| + | *the fMRI contains low tissue contrast, making automated intensity-based registration difficult. | ||
*the two images often have strong differences in voxel sizes and voxel anisotropy. If the orientation of the highest resolution is not the same in both images, finding a good match can be difficult. | *the two images often have strong differences in voxel sizes and voxel anisotropy. If the orientation of the highest resolution is not the same in both images, finding a good match can be difficult. | ||
*there may be widespread and extensive pathology (e.g stroke, tumor) that might affect the registration if its contrast is different in the baseline and structural reference scan | *there may be widespread and extensive pathology (e.g stroke, tumor) that might affect the registration if its contrast is different in the baseline and structural reference scan | ||
=== Discussion: Key Strategies === | === Discussion: Key Strategies === | ||
| − | |||
| − | |||
| − | |||
*masking is likely necessary to obtain good results. | *masking is likely necessary to obtain good results. | ||
| − | *in this example the initial alignment of the two scans is | + | *in this example the initial alignment of the two scans is not excessive. |
| − | |||
*because speed is not that critical, we increase the sampling rate from the default 2% to 15%. | *because speed is not that critical, we increase the sampling rate from the default 2% to 15%. | ||
*we also expect larger differences in scale & distortion than with regular structural scane: so we significantly (2x-3x) increase the expected values for scale and skew from the defaults. | *we also expect larger differences in scale & distortion than with regular structural scane: so we significantly (2x-3x) increase the expected values for scale and skew from the defaults. | ||
Revision as of 21:18, 3 February 2010
Home < Projects:RegistrationLibrary:RegLib C09Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
Slicer Registration Library Exampe #9: Functional MRI aligned with structural reference MRI
RegLib 09: T1 SPGR RegLib 09: fMR
| this is the fixed reference image. All images are aligned into this space | this is the moving image. The transform is calculated by matching this to the reference image
| |
| 0.46 x 0.46 x 3.0 mm axial 512 x 512 x 46 RAS |
1.0 x 1.0 x 3.3 mm axial oblique 256 x 256 x 36 RAS |
Objective / Background
This is a typical example of fMRI pre-processing. Goal is to align the fMRI image with a structural scan that provides accuracte anatomical reference. The fMRI contains acquisition-related distortion and low contrast to discern much anatomical detail. We also have pathology (stroke) with variable contrast across different MRI protocols.
Keywords
MRI, brain, head, intra-subject, fMRI
Input Data
Registration Results
Download
- download entire package (Data,Presets,Tutorial, Solution, zip file 33.7 MB) (to be added: tutorial + presets)
- Presets
- Tutorial only
- Image Data only
Discussion: Registration Challenges
- the fMRI contains acquisition-related distortions that can make automated registration difficult.
- the fMRI contains low tissue contrast, making automated intensity-based registration difficult.
- the two images often have strong differences in voxel sizes and voxel anisotropy. If the orientation of the highest resolution is not the same in both images, finding a good match can be difficult.
- there may be widespread and extensive pathology (e.g stroke, tumor) that might affect the registration if its contrast is different in the baseline and structural reference scan
Discussion: Key Strategies
- masking is likely necessary to obtain good results.
- in this example the initial alignment of the two scans is not excessive.
- because speed is not that critical, we increase the sampling rate from the default 2% to 15%.
- we also expect larger differences in scale & distortion than with regular structural scane: so we significantly (2x-3x) increase the expected values for scale and skew from the defaults.
- a good affine alignment is important before proceeding to non-rigid alignment to further correct for distortions.