Difference between revisions of "Projects:RegistrationLibrary:RegLib C11"
From NAMIC Wiki
(Created page with 'Back to ARRA main page <br> Back to Registration main page <br> [[Projects:RegistrationDocumentation:UseCaseInv…') |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| (25 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | ==Slicer Registration Library | + | ==<small>v3.6.1</small> [[Image:Slicer3-6Announcement-v1.png|150px]] Slicer Registration Library Case #11: <br>Co-registration of two anatomic brain atlases for merging of structure labels == |
| − | + | === Input === | |
| − | {| style="color:#bbbbbb | + | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" |
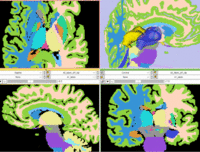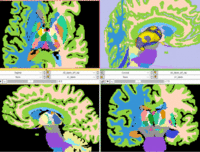
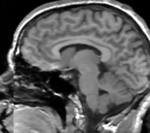
| − | |[[Image: | + | |[[Image:RegLib_C11_A1_gray.png|150px|lleft|this is the fixed reference image. All images are aligned into this space]] |
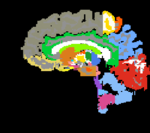
| − | |[[Image: | + | |[[Image:RegLib_C11_A1_label.png|150px|lleft|this is the fixed target, a label-map in the same space as the fixed reference. The registration target is derived from this image]] |
| − | + | |[[Image:RegArrow_Affine.png|100px|lleft]] | |
| − | | | + | |[[Image:RegLib_C11_A0_gray.png|150px|lleft|this is the moving image, but the transform is calculated by matching surfaces derived from the labelmaps]] |
| − | [[Image: | + | |[[Image:RegLib_C11_A0_label.png|150px|lleft|this is the moving image to which the calculated transform is applied. The model surfaces used to calculate the transform are derived from this image]] |
| − | [[Image: | ||
| − | |||
| − | |||
| − | |[[Image: | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | |Target Atlas T1 |
| + | |Target Atlas Labelmap | ||
| | | | ||
| − | | | + | |Moving Atlas T1 |
| + | |Moving Atlas Labelmap | ||
|} | |} | ||
| + | |||
| + | === Modules === | ||
| + | '''Slicer 3.6.1''' recommended modules: [https://www.slicer.org/wiki/Modules:PythonSurfaceICPRegistration-Documentation-3.6 ''Surface Registration'''] | ||
| + | |||
| + | ===Download === | ||
| + | *Data | ||
| + | **[[Media:RegLib_C11_AtlasMerge_TutorialSet.zip|download entire '''tutorial package''' <small> (Original Data, Intermediate Results, Solution, zip file 74 MB) </small>]] | ||
| + | **[[Media:RegLib_C11_AtlasMerge_OldAtlas.zip|download '''old atlas dataset''' <small> (MRI+labelmap,thalamic nuclei models, list of labels+names), zip file 5.4 MB) </small>]] | ||
| + | **[[Media:RegLib_C11_AtlasMerge_NewAtlas.zip|download '''new atlas dataset''' <small> (MRI T1+T2, labelmap, thalamus model), zip file 126MB) </small>]] | ||
| + | **[[Media:RegLib_C11_AtlasMerge_NewAtlasMerged.zip|download '''result merged new atlas dataset''' <small> (MRI T1+T2, new merged labelmaps (2x) incl. thalamic nuclei, colormap file with label names), zip file 132 MB) </small>]] | ||
| + | **[[Media:RegLib_C11_Resampled.zip|download '''resampled atlas dataset''' <small> (T1 only resampled to 1mm isotropic, padded to 256 , labelmap reoriented as sagittal), zip file 9 MB) </small>]] | ||
| + | *Documentation | ||
| + | **[[Media:RegLib_C11_Tutorial_AtlasMerging.ppt|download full '''step-by-step tutorial'''<small> (PowerPoint file, 2.5 MB) </small>]] | ||
| + | **[[Media:RegLib_C11_Tutorial_AtlasMerging.pdf|download full '''step-by-step tutorial'''<small> (PDF file, 2.8 MB) </small>]] | ||
===Objective / Background === | ===Objective / Background === | ||
| − | This is an example of | + | This is an example of inter-subject registration via surface matching. The structures of interest are a small subset of the entire image, hence registration is not driven by image intensities but rather two model surfaces derived from the labelmaps. |
| + | |||
=== Keywords === | === Keywords === | ||
| − | MRI, brain, head, inter-subject, | + | MRI, brain, head, inter-subject, atlas to atlas, surface-based registration, thalamic nuclei |
===Input Data=== | ===Input Data=== | ||
| − | * | + | *fixed : T1w coronal, 1mm isotropic. Called ''A1_gray''; 1mm isotropic, 256 x 256 x 256 |
| − | * | + | *fixed : labelmap , aligned with above. Called ''A1_label''; 1mm isotropic, 256 x 256 x 256 |
| + | *moving: T1w coronal, 0.9 inplane, 1.5mm coronal slices. Called ''A0_gray''; 0.9375 x 0.9375 x 1.5mm isotropic, 256 x 256 x 159 | ||
| + | *moving: labelmap , aligned with above. Called ''A0_label''; 0.9375 x 0.9375 x 1.5mm isotropic, 256 x 256 x 159 | ||
| + | |||
=== Methods === | === Methods === | ||
| − | # | + | #Visualize & browse A0 data: determine label range of thalamic nuclei labels in A0_label: 500-526 |
| − | # | + | #Visualize & browse A1 data: determine label range of thalamus lables in A1_label: 10 and 49 |
| − | # | + | #Build label mask of thalamus for A0: '''Editor''' module |
| − | :: | + | ##Create Labelmap From”: ''A0_labels'' |
| − | :: | + | ##Select Labelmap to Edit: select the newly created “A0_labels-label” and then select “Rename”. Rename the new volume to ''A0_thalamus'' |
| − | :: | + | ##From the icon panel, select the “Threshold Icon” |
| − | :: | + | ##Threshold range: enter 500 and 526 |
| − | + | #change ''A1_labels'': change label 10 to 49; '''Editor''' module | |
| − | + | ##Select Labelmap to Edit: ''Aa_labels'' | |
| − | : | + | ##Label field: enter 49 |
| − | :: | + | ##Change Island Icon; left click on area with left thalamus label 10 |
| − | :: | + | #Build label mask of thalamus for A1: '''Editor''' module |
| − | :: | + | ##Create Labelmap From”: ''A1_labels'' |
| − | :: | + | ##Select Labelmap to Edit: select the newly created “A1_labels-label” and then select “Rename”. Rename the new volume to ''A1_thalamus'' |
| − | :: | + | ##From the icon panel, select the “Threshold Icon” |
| − | :: | + | ##Threshold range: enter 49 and 49 |
| − | :: | + | #Smooth A1 thalamus labelmap: '''Filtering / Denoising / Median Filter''' |
| − | # | + | ##Neighborhood Size: (default) 1,1,1 |
| − | # | + | ##Input Volume: A1_thalamus |
| − | :: | + | ##Output Volume: A1_thalamus |
| − | : | + | #Build thalamus surface model of A0: '''Model Maker''' module |
| − | :: | + | ##Input Volume: ''A0_thalamus'' |
| − | :: | + | ##Model Name: ''A0_ThalamusModel'' |
| + | ##Labels: 1 | ||
| + | ##Smooth: 50 | ||
| + | ##Decimate: 0.25 | ||
| + | ##Split Normals: no | ||
| + | ##Point Normals: yes | ||
| + | ##Save Intermediate Models: no | ||
| + | #Build thalamus surface model of A1 ('''Model Maker''' module as above, with ''A1_thalamus'' as input and ''A1_ThalamusModel'' as output. | ||
| + | #Co-register thalamus surfaces: '''Python Surface ICP Registration''' module. Settings: | ||
| + | ##Landmark transform mode: Affine | ||
| + | ##Mean Distance Mode: RMS | ||
| + | ##Maximum Number of Iterations: 500 | ||
| + | ##Maximum Nmber of Landmarks: 200 | ||
| + | ##Start by matching centroids: yes | ||
| + | ##maximum mean distance: 0.01 | ||
| + | ##Initial Transform: none | ||
| + | ##Input Surface: ''A0_ThalamusModel'' | ||
| + | ##Target Surface: ''A1_ThalamusModel'' | ||
| + | ##Output Surface: none | ||
| + | ##Output Transform: ''Xform_A0Affine_ICP'' | ||
| + | # Apply Registration Xform to labelmap: '''Resample Scalar/Vector/DWI Volume''' module. Settings: | ||
| + | ##Input Volume : ''A0_labels'' | ||
| + | ##Reference Volume : ''A1_labels'' | ||
| + | ##Output Volume : “Create New Volume”, rename to “A0_labels_aff” | ||
| + | ##Transform Node: ''Xform_A0Affine_ICP'' | ||
| + | ##Interpolation Type: “nn” | ||
| + | ##Change datatype of new volume ''A0_labels_aff'' to labelmap ('''Volumes''' module) | ||
| + | # Apply Registration Xform to thalamus mask: '''Resample Scalar/Vector/DWI Volume''' module. Settings: | ||
| + | ##Input Volume : ''A0_thalamus'' | ||
| + | ##Reference Volume : ''A1_labels'' | ||
| + | ##Output Volume : “Create New Volume”, rename to ''A0_thalamus_aff'' | ||
| + | ##Transform Node: ''Xform_A0Affine_ICP'' | ||
| + | ##Interpolation Type: “nn” | ||
| + | ##Change datatype of new volume ''A0_labels_aff'' to labelmap ('''Volumes''' module) | ||
| + | #Mask new labelmap with ''A0_thalamus_aff'': '''Mask Image''' module | ||
| + | ##Input Volume : ''A0_labels_aff'' | ||
| + | ##Mask Volume : ''A0_thalamus_aff'' | ||
| + | ##Masked Volume: ''A0_labels aff'' | ||
| + | ##Mask new labelmap with A1_thalamus: '''Mask Image''' module | ||
| + | ##Input Volume : ''A0_labels_aff'' | ||
| + | ##Mask Volume : ''A1_thalamus_aff'' | ||
| + | ##Masked Volume: New volume, rename to: ''A0_labels aff_clip'' | ||
| + | #Typecast A1_labelmap: '''Cast Image''' module | ||
| + | ##Input Volume: ''A1_label'' | ||
| + | ##Output Volume: ''A1_label'' | ||
| + | ##Output Type: “short” | ||
| + | #Merge Labelmaps:'''Image Label Combine''' module | ||
| + | ##Input Label Map A: ''A0_label_aff_clip'' | ||
| + | ##Input Label Map B: ''A1_labels'' | ||
| + | ##Output Label Map: “Create New Volume”, rename to ''A1_labels_merged'' | ||
| + | ##First label overwrites second: yes | ||
=== Registration Results=== | === Registration Results=== | ||
{| style="color:#bbbbbb; background-color:#333333;" cellpadding="10" cellspacing="0" border="0" | {| style="color:#bbbbbb; background-color:#333333;" cellpadding="10" cellspacing="0" border="0" | ||
| − | |[[Image: | + | |[[Image:RegLib_C11_AnimGif2.gif|200px|left|unregistered]] [[Image:RegLib_C11_AnimGif1.gif|200px|left|after ICP Surface Registration + Clipping]] |
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Discussion: Registration Challenges === | === Discussion: Registration Challenges === | ||
| − | *Because the | + | *Because the structures of interest are a very small subset of the image without distinct grayscale contrast |
| − | * | + | *the two atlases represent different anatomies and hence some residual misalignment is inevitable |
| − | * | + | *the two labelmaps have different resolutions and different smoothness of structure outlines. Some need filtering to remove spurious surface details that would distract the registration algorithm |
=== Discussion: Key Strategies === | === Discussion: Key Strategies === | ||
| − | *Because of the | + | *Because the structures of interest are a very small subset of the image without distinct grayscale contrast, we co-register surfaces rather than intensity volumes |
| − | + | ||
| − | |||
| − | |||
| − | |||
=== Acknowledgments === | === Acknowledgments === | ||
| − | *dataset provided by | + | *dataset provided by Ron Kikinis, M.D. and Florin Talos, M.D. |
Latest revision as of 17:17, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C11Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
v3.6.1  Slicer Registration Library Case #11:
Slicer Registration Library Case #11:
Co-registration of two anatomic brain atlases for merging of structure labels
Input
|
|
|
|
|
| Target Atlas T1 | Target Atlas Labelmap | Moving Atlas T1 | Moving Atlas Labelmap |
Modules
'Slicer 3.6.1 recommended modules: Surface Registration
Download
- Data
- download entire tutorial package (Original Data, Intermediate Results, Solution, zip file 74 MB)
- download old atlas dataset (MRI+labelmap,thalamic nuclei models, list of labels+names), zip file 5.4 MB)
- download new atlas dataset (MRI T1+T2, labelmap, thalamus model), zip file 126MB)
- download result merged new atlas dataset (MRI T1+T2, new merged labelmaps (2x) incl. thalamic nuclei, colormap file with label names), zip file 132 MB)
- download resampled atlas dataset (T1 only resampled to 1mm isotropic, padded to 256 , labelmap reoriented as sagittal), zip file 9 MB)
- Documentation
Objective / Background
This is an example of inter-subject registration via surface matching. The structures of interest are a small subset of the entire image, hence registration is not driven by image intensities but rather two model surfaces derived from the labelmaps.
Keywords
MRI, brain, head, inter-subject, atlas to atlas, surface-based registration, thalamic nuclei
Input Data
- fixed : T1w coronal, 1mm isotropic. Called A1_gray; 1mm isotropic, 256 x 256 x 256
- fixed : labelmap , aligned with above. Called A1_label; 1mm isotropic, 256 x 256 x 256
- moving: T1w coronal, 0.9 inplane, 1.5mm coronal slices. Called A0_gray; 0.9375 x 0.9375 x 1.5mm isotropic, 256 x 256 x 159
- moving: labelmap , aligned with above. Called A0_label; 0.9375 x 0.9375 x 1.5mm isotropic, 256 x 256 x 159
Methods
- Visualize & browse A0 data: determine label range of thalamic nuclei labels in A0_label: 500-526
- Visualize & browse A1 data: determine label range of thalamus lables in A1_label: 10 and 49
- Build label mask of thalamus for A0: Editor module
- Create Labelmap From”: A0_labels
- Select Labelmap to Edit: select the newly created “A0_labels-label” and then select “Rename”. Rename the new volume to A0_thalamus
- From the icon panel, select the “Threshold Icon”
- Threshold range: enter 500 and 526
- change A1_labels: change label 10 to 49; Editor module
- Select Labelmap to Edit: Aa_labels
- Label field: enter 49
- Change Island Icon; left click on area with left thalamus label 10
- Build label mask of thalamus for A1: Editor module
- Create Labelmap From”: A1_labels
- Select Labelmap to Edit: select the newly created “A1_labels-label” and then select “Rename”. Rename the new volume to A1_thalamus
- From the icon panel, select the “Threshold Icon”
- Threshold range: enter 49 and 49
- Smooth A1 thalamus labelmap: Filtering / Denoising / Median Filter
- Neighborhood Size: (default) 1,1,1
- Input Volume: A1_thalamus
- Output Volume: A1_thalamus
- Build thalamus surface model of A0: Model Maker module
- Input Volume: A0_thalamus
- Model Name: A0_ThalamusModel
- Labels: 1
- Smooth: 50
- Decimate: 0.25
- Split Normals: no
- Point Normals: yes
- Save Intermediate Models: no
- Build thalamus surface model of A1 (Model Maker module as above, with A1_thalamus as input and A1_ThalamusModel as output.
- Co-register thalamus surfaces: Python Surface ICP Registration module. Settings:
- Landmark transform mode: Affine
- Mean Distance Mode: RMS
- Maximum Number of Iterations: 500
- Maximum Nmber of Landmarks: 200
- Start by matching centroids: yes
- maximum mean distance: 0.01
- Initial Transform: none
- Input Surface: A0_ThalamusModel
- Target Surface: A1_ThalamusModel
- Output Surface: none
- Output Transform: Xform_A0Affine_ICP
- Apply Registration Xform to labelmap: Resample Scalar/Vector/DWI Volume module. Settings:
- Input Volume : A0_labels
- Reference Volume : A1_labels
- Output Volume : “Create New Volume”, rename to “A0_labels_aff”
- Transform Node: Xform_A0Affine_ICP
- Interpolation Type: “nn”
- Change datatype of new volume A0_labels_aff to labelmap (Volumes module)
- Apply Registration Xform to thalamus mask: Resample Scalar/Vector/DWI Volume module. Settings:
- Input Volume : A0_thalamus
- Reference Volume : A1_labels
- Output Volume : “Create New Volume”, rename to A0_thalamus_aff
- Transform Node: Xform_A0Affine_ICP
- Interpolation Type: “nn”
- Change datatype of new volume A0_labels_aff to labelmap (Volumes module)
- Mask new labelmap with A0_thalamus_aff: Mask Image module
- Input Volume : A0_labels_aff
- Mask Volume : A0_thalamus_aff
- Masked Volume: A0_labels aff
- Mask new labelmap with A1_thalamus: Mask Image module
- Input Volume : A0_labels_aff
- Mask Volume : A1_thalamus_aff
- Masked Volume: New volume, rename to: A0_labels aff_clip
- Typecast A1_labelmap: Cast Image module
- Input Volume: A1_label
- Output Volume: A1_label
- Output Type: “short”
- Merge Labelmaps:Image Label Combine module
- Input Label Map A: A0_label_aff_clip
- Input Label Map B: A1_labels
- Output Label Map: “Create New Volume”, rename to A1_labels_merged
- First label overwrites second: yes
Registration Results
Discussion: Registration Challenges
- Because the structures of interest are a very small subset of the image without distinct grayscale contrast
- the two atlases represent different anatomies and hence some residual misalignment is inevitable
- the two labelmaps have different resolutions and different smoothness of structure outlines. Some need filtering to remove spurious surface details that would distract the registration algorithm
Discussion: Key Strategies
- Because the structures of interest are a very small subset of the image without distinct grayscale contrast, we co-register surfaces rather than intensity volumes
Acknowledgments
- dataset provided by Ron Kikinis, M.D. and Florin Talos, M.D.