Difference between revisions of "Projects:RegistrationLibrary:RegLib C13"
From NAMIC Wiki
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | ==<small> | + | == <small>updated for '''v4.1'''</small> [[Image:Slicer4_RegLibLogo.png|150px]] <br> Slicer Registration Library Case #13: Liver Tumor Cryoablation 2 == |
=== Input === | === Input === | ||
| Line 13: | Line 13: | ||
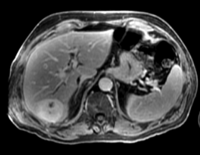
|[[Image:RegLib_C13_LiverTumor2_MRIpre_T2.png|200px|lleft|this is the moving image. The transform is calculated by matching this to the reference image]] | |[[Image:RegLib_C13_LiverTumor2_MRIpre_T2.png|200px|lleft|this is the moving image. The transform is calculated by matching this to the reference image]] | ||
|- | |- | ||
| − | |fixed image/target | + | |fixed image/target: CT post |
| | | | ||
| − | |moving image | + | |moving image 1: CT intra |
| − | |moving image | + | |moving image 2: MRI T1 pre |
| − | |moving image | + | |moving image 3: MRI T2 pre |
|} | |} | ||
===Objective / Background === | ===Objective / Background === | ||
| − | We seek to align | + | We seek to align/fuse pre-operative MRI with intra- and post-operative CT. |
=== Keywords === | === Keywords === | ||
| Line 27: | Line 27: | ||
===Input Data=== | ===Input Data=== | ||
| − | *reference/fixed : 0.57 x 0.57 x 5 , 512 x 512 x 37 voxels | + | *CT post reference/fixed : 0.57 x 0.57 x 5 , 512 x 512 x 37 voxels |
| − | * | + | *CT pre: 0.57 x 0.57 x 5 mm, 512 x 512 x 33 voxels |
| − | * | + | *MRI T1 pre: 1.56 x 1.56 x 2.5 mm, 256 x 256 x 88 |
| − | * | + | *MRI T2 pre: 1.56 x 1.56 x 6 mm, 256 x 256 x 36 |
| − | |||
| − | |||
===Download === | ===Download === | ||
| Line 43: | Line 41: | ||
=== Procedure === | === Procedure === | ||
*'''Phase I: register CTpre - CTpost''' | *'''Phase I: register CTpre - CTpost''' | ||
| + | #this registration is fairly straightforward, since FOV, contrast and initial orientation of the two scans are very similar. We perform a linear + nonrigid registration without any masking | ||
#open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] | ||
##''Fixed Image Volume'': CTpost | ##''Fixed Image Volume'': CTpost | ||
| Line 55: | Line 54: | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
##click: Apply; runtime ~ 1 min (MacPro QuadCore 2.4GHz) | ##click: Apply; runtime ~ 1 min (MacPro QuadCore 2.4GHz) | ||
| + | *'''Phase II: Create Masks''' | ||
| + | #for the MR-CT registration we have a big difference in the FOV as well as contrast and amount of anisotropy. We need to mask the liver as region of interest to stabilize the registration; otherwise the clipped FOV will cause the moving image to distort. | ||
| + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/Editor Editor module] | ||
| + | #select "CTpost" as the master volume , a new CTpost_label volume will be created. | ||
| + | #select the ''Brush'' tool and manually cover the liver and adjacent areas in all axial slices. This need not be accurate, the goal is to exclude the contralateral side where the FOV is clipped. To skip this step you may load the masks provided with the example data. | ||
| + | #repeat and create a similar mask for the MRI-T2. | ||
| + | #save results thus far | ||
| + | *'''Phase III: Nonrigid Registration MR-CT''' | ||
| + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] | ||
| + | ##''Fixed Image Volume'': CTpost | ||
| + | ##''Moving Image Volume'': MRI_T2 | ||
| + | ##Output Settings: | ||
| + | ###''Slicer BSpline Transform": none | ||
| + | ###''Slicer Linear Transform'': create new transform, rename to "Xf2_T2-CTpost_Affine.tfm" | ||
| + | ###''Output Image Volume'': create new volume, rename to "T2_Xf2" (we use this for validation only) | ||
| + | ##''Registration Phases'': check boxes for ''Rigid'', "Rigid+Scale", "Affine" and "BSpline" | ||
| + | ##''Main Parameters'': | ||
| + | ##''Mask Option'': select ''ROI'' button | ||
| + | ###''ROI Masking input fixed'': select "CTpost_label" created in phase II above | ||
| + | ###''ROI Masking input moving'': select "MRI-T2-label.nrrd" created in phase II above | ||
| + | ##Leave all other settings at default | ||
| + | ##click: Apply | ||
| Line 62: | Line 83: | ||
*contrast enhancement and pathology and treatment changes cause additional differences in image content | *contrast enhancement and pathology and treatment changes cause additional differences in image content | ||
*we have strongly anisotropic voxel sizes with much less through-plane resolution | *we have strongly anisotropic voxel sizes with much less through-plane resolution | ||
| + | |||
| + | === Registration Results=== | ||
Revision as of 20:45, 10 May 2012
Home < Projects:RegistrationLibrary:RegLib C13Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
updated for v4.1 
Slicer Registration Library Case #13: Liver Tumor Cryoablation 2
Input
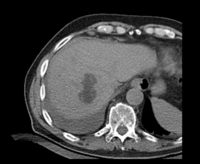
|
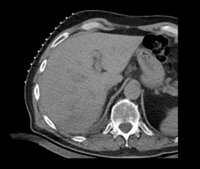
|
|
|
|
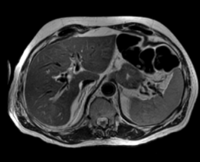
| fixed image/target: CT post | moving image 1: CT intra | moving image 2: MRI T1 pre | moving image 3: MRI T2 pre |
Objective / Background
We seek to align/fuse pre-operative MRI with intra- and post-operative CT.
Keywords
MRI, CT, IGT, intra-operative, liver, cryoablation, change detection, non-rigid registration
Input Data
- CT post reference/fixed : 0.57 x 0.57 x 5 , 512 x 512 x 37 voxels
- CT pre: 0.57 x 0.57 x 5 mm, 512 x 512 x 33 voxels
- MRI T1 pre: 1.56 x 1.56 x 2.5 mm, 256 x 256 x 88
- MRI T2 pre: 1.56 x 1.56 x 6 mm, 256 x 256 x 36
Download
Procedure
- Phase I: register CTpre - CTpost
- this registration is fairly straightforward, since FOV, contrast and initial orientation of the two scans are very similar. We perform a linear + nonrigid registration without any masking
- open the General Registration (BRAINS) module
- Fixed Image Volume: CTpost
- Moving Image Volume: CTpre
- Output Settings:
- Slicer BSpline Transform": create new transform, rename to "Xf1_Ctpre-CTpost_BSpline"
- Slicer Linear Transform: none
- Output Image Volume: create new volume, rename to "CTpre_Xf1"
- Registration Phases: check boxes for Rigid , Rigid+Scale , Affine and "BSpline"
- Main Parameters:
- B-Spline Grid Size: 7,7,3
- Leave all other settings at default
- click: Apply; runtime ~ 1 min (MacPro QuadCore 2.4GHz)
- Phase II: Create Masks
- for the MR-CT registration we have a big difference in the FOV as well as contrast and amount of anisotropy. We need to mask the liver as region of interest to stabilize the registration; otherwise the clipped FOV will cause the moving image to distort.
- open the Editor module
- select "CTpost" as the master volume , a new CTpost_label volume will be created.
- select the Brush tool and manually cover the liver and adjacent areas in all axial slices. This need not be accurate, the goal is to exclude the contralateral side where the FOV is clipped. To skip this step you may load the masks provided with the example data.
- repeat and create a similar mask for the MRI-T2.
- save results thus far
- Phase III: Nonrigid Registration MR-CT
- open the General Registration (BRAINS) module
- Fixed Image Volume: CTpost
- Moving Image Volume: MRI_T2
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create new transform, rename to "Xf2_T2-CTpost_Affine.tfm"
- Output Image Volume: create new volume, rename to "T2_Xf2" (we use this for validation only)
- Registration Phases: check boxes for Rigid, "Rigid+Scale", "Affine" and "BSpline"
- Main Parameters:
- Mask Option: select ROI button
- ROI Masking input fixed: select "CTpost_label" created in phase II above
- ROI Masking input moving: select "MRI-T2-label.nrrd" created in phase II above
- Leave all other settings at default
- click: Apply
Discussion: Registration Challenges
- large differences in FOV
- strong differences in image contrast between MRI & CT
- contrast enhancement and pathology and treatment changes cause additional differences in image content
- we have strongly anisotropic voxel sizes with much less through-plane resolution