Difference between revisions of "Projects:RegistrationLibrary:RegLib C18"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (14 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | = <small>v3.6.1</small> [[Image:Slicer3-6Announcement-v1.png|150px]] Slicer Registration Library Case 18: <br> | + | = <small>v3.6.1</small> [[Image:Slicer3-6Announcement-v1.png|150px]] Slicer Registration Library Case 18: <br> Serial Multi-contrast MRI in Aging Study = |
=== Input === | === Input === | ||
| − | {| style="color:#bbbbbb; " cellpadding=" | + | {| style="color:#bbbbbb; " cellpadding="2" cellspacing="0" border="0" |
| − | |[[Image: | + | |[[Image:RegLib_C18_Thumb1.png|100px|lleft]] |
| − | |||
|[[Image:RegArrow_Affine.png|70px|lleft]] | |[[Image:RegArrow_Affine.png|70px|lleft]] | ||
| − | |[[Image: | + | |[[Image:RegLib_C18_Thumb2.png|100px|lleft]] |
| + | |[[Image:RegLib_C18_Thumb3.png|100px|lleft]] | ||
|- | |- | ||
| − | |exam 1: | + | |exam 1: T1 |
| + | | | ||
| + | |exam 1: FLAIR | ||
|exam 1: T2 | |exam 1: T2 | ||
| − | |||
| − | |||
|- | |- | ||
|[[Image:RegArrow_AffineVert.png|70px|lleft]] | |[[Image:RegArrow_AffineVert.png|70px|lleft]] | ||
| Line 21: | Line 21: | ||
| | | | ||
|- | |- | ||
| − | |[[Image: | + | |[[Image:RegLib_C18_Thumb4.png|100px|lleft]] |
| − | |||
|[[Image:RegArrow_Affine.png|70px|lleft]] | |[[Image:RegArrow_Affine.png|70px|lleft]] | ||
| − | |[[Image: | + | |[[Image:RegLib_C18_Thumb5.png|100px|lleft]] |
| + | |[[Image:RegLib_C18_Thumb6.png|100px|lleft]] | ||
|- | |- | ||
| − | |exam 2: | + | |exam 2: T1 |
| + | | | ||
| + | |exam 2: FLAIR | ||
|exam 2: T2 | |exam 2: T2 | ||
| − | |||
| − | |||
|} | |} | ||
=== Modules === | === Modules === | ||
| − | *'''Slicer 3.6.1 recommended modules: [ | + | *'''Slicer 3.6.1 recommended modules: [https://www.slicer.org/wiki/Modules:BRAINSFit BrainsFit], [https://www.slicer.org/wiki/Modules:RegisterImages-Documentation-3.6 Expert Automated Registration] |
===Objective / Background === | ===Objective / Background === | ||
| − | This scenario occurs in many forms whenever we wish to assess change in a series of multi-contrast MRI. The follow-up scan(s) are to be aligned with the baseline, but also the different series within each exam need to be co-registered, since the subject may have moved between acquisitions. Hence we have a set of nested registrations. This particular exam features | + | This scenario occurs in many forms whenever we wish to assess change in a series of multi-contrast MRI. The follow-up scan(s) are to be aligned with the baseline, but also the different series within each exam need to be co-registered, since the subject may have moved between acquisitions. Hence we have a set of nested registrations. This particular exam features T1-, FLAIR and T2-weighted scans. |
| + | |||
=== Download === | === Download === | ||
*Data: | *Data: | ||
| − | **[[Media:RegLib_C18_Data.zip|'''Registration Library Case | + | **[[Media:RegLib_C18_Data.zip|'''Registration Library Case 18: Longitudinal multi-contrast series (T1,T2,FLAIR)''' <small> (Data,Presets, Solution, zip file 49 MB) </small>]] |
*Presets | *Presets | ||
| − | **[[Media: | + | **[[Media:RegLib_C18_Registration_Presets.mrml|Registration Presets <small> (mrml file , 12kB) </small>]] |
**[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]]</small> | **[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]]</small> | ||
*Documentation | *Documentation | ||
| Line 72: | Line 73: | ||
*'''Load & Center''' | *'''Load & Center''' | ||
#Load example dataset via OpenScene... | #Load example dataset via OpenScene... | ||
| + | #Note that when you load the data as a scene, the image volumes have deliberately '''not''' been centered to demonstrate the effect of different image origins. Hence the result transforms provided will also not look right until the centering below is performed: | ||
#Go to the ''Data'' module. You should see 6 images (e1_PD, e1_T2 etc.) and 3 solution transforms (Xform_...) | #Go to the ''Data'' module. You should see 6 images (e1_PD, e1_T2 etc.) and 3 solution transforms (Xform_...) | ||
#Set background view to e1_PD and foreground to e2_PD. Toggle to see misalignment | #Set background view to e1_PD and foreground to e2_PD. Toggle to see misalignment | ||
| Line 116: | Line 118: | ||
##Registration Parameters: increase the ''Number of Samples'' field to 200,000 | ##Registration Parameters: increase the ''Number of Samples'' field to 200,000 | ||
##Leave all other settings at defaults & Click: ''Apply''. Registration should complete within ~ 30 seconds | ##Leave all other settings at defaults & Click: ''Apply''. Registration should complete within ~ 30 seconds | ||
| + | *'''Align Exam 2:''' | ||
| + | **repeat the above steps for exam 2, i.e. align e2_FLAIR and e2_T2 with e2_T1 | ||
| + | **the above issue with the FLAIR-T1 residual does not arise for the follow-up exam, hence direct registration (unmasked) is possible | ||
| + | **Registration presets: BRAINSFit see Xf3_ and Xf4_ for FLAIR and T2, respectively | ||
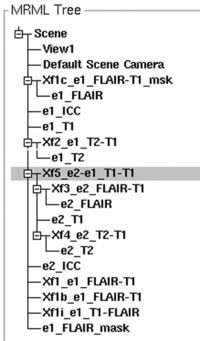
| + | [[Image:RegLib_C18_RegHierarchy.png|200px|right| the MRML node tree shows the hierarchy of multiple transfoms]] the MRML node tree shows the hierarchy of multiple transfoms. | ||
*'''Align Exam 2-1:''' | *'''Align Exam 2-1:''' | ||
| − | #we | + | #finally we align the T1 references of exam 1 and 2, i.e. we align e2_T1 with e1_T1: |
| − | + | #Registration presets: BRAINSFit see Xf5_ | |
| − | + | #parameters: defaults except increase sample rate to 200,000. | |
| − | + | #here we have genuine masks for both images: ''Control of Mask Processing Tab'': | |
| − | + | ##Control of Mask Processing tab: check "ROI" box, | |
| − | + | ##Input Fixed Mask, select "e1_ICC", | |
| − | # | + | ##Input Moving Mask, select "e2_ICC" |
| − | # | + | ##Leave all other settings at defaults & Click: ''Apply''. Registration should complete within ~ 20 seconds |
| − | ## | + | *'''Combine Transforms:''' |
| − | ## | + | #to align all images in a common space, we combine a hierarchy of transforms, i.e. the e2_FLAIR undergoes both the Xf3_e2_FLAIR-T1 and the Xf4_e2-e1: See image on the right for what the final hierarchy should look like. |
| − | ## | + | #right-click on the nested transforms and select ''Harden Transforms'' from the pulldown menu. Rename the hardened transforms to indicate the direct alignment with e1_T1. |
| − | + | *''Resample'' | |
| − | ##Leave all other settings at defaults | + | #resample all images with their respective transforms, using the ''Filtering -> Resample Scalar/Vector/DWI Volume'' module. |
| − | + | #within the module, select "ws" as interpolation type | |
| − | + | *''Nonrigid extension'' | |
| − | + | #there is some residual distortion in the alignment of e2_T1 to e1_T1. Optionally one can address this via another separate nonrigid BSpline registration. However combining the Affine with the BSpline is not straightforward (as of v.3.6.1), to do this follow this outline: | |
| − | + | ##resample e2_T1 according to Xf5 : e2_T1_Xf5 | |
| − | # | + | ##Perform a BSpline ''only'' alignment : Xf6_e2-e1_BSpline that aligns e2_T1_Xf5 with e1_T1. Grid size 3-5. |
| − | + | ##Save resulting Xform | |
| − | + | ##Open Xform file and other saved affine transforms in Text Editor | |
| − | # | + | ##Paste the affine portion at the bottom of the BSpline and save under new name. |
| − | |||
| − | |||
| − | # | ||
| − | ## | ||
| − | ## | ||
| − | |||
| − | |||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
| + | [[Image:RegLib_C18_AGif_e1.gif|300px|Exam 1 co-registered]] Exam 1 co-registered: FLAIR and T2 aligned with T1 <br><br> | ||
| + | [[Image:RegLib_C18_AGif_e2.gif|300px|Exam 2 co-registered]] Exam 1 co-registered: FLAIR and T2 aligned with T1 <br><br> | ||
| + | [[Image:RegLib_C18_AGif_e2-e1.gif|300px|Exam 2 aligned to Exam 1]] Exam 2 aligned to Exam 1 (showing FLAIR and T2 of exam 2 also) <br> | ||
Latest revision as of 18:07, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C18Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.1  Slicer Registration Library Case 18:
Slicer Registration Library Case 18:
Serial Multi-contrast MRI in Aging Study
Input
|
|
|
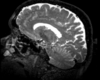
|
| exam 1: T1 | exam 1: FLAIR | exam 1: T2 | |

|
|||
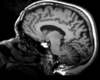
|

|
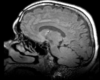
|
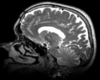
|
| exam 2: T1 | exam 2: FLAIR | exam 2: T2 |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit, Expert Automated Registration
Objective / Background
This scenario occurs in many forms whenever we wish to assess change in a series of multi-contrast MRI. The follow-up scan(s) are to be aligned with the baseline, but also the different series within each exam need to be co-registered, since the subject may have moved between acquisitions. Hence we have a set of nested registrations. This particular exam features T1-, FLAIR and T2-weighted scans.
Download
- Data:
- Presets
- Documentation
- see Methods section below for step-by-step instructions
Keywords
MRI, brain, head, intra-subject, multiple sclerosis, MS, multi-contrast, change assessment, dual echo, nested registration
Input Data
- reference/fixed : PD.1 baseline exam , 0.9375 x 0.9375 x 3 mm voxel size, axial acquisition, RAS orientation.
- fixed T2.1 baseline exam , 0.9375 x 0.9375 x 3 mm voxel size, axial acquisition, RAS orientation. -> (aligned with PD.1, not used for registering)
- moving: T1.1 (GdDTPA contrast-enhanced scan) baseline exam 0.9375 x 0.9375 x 5 mm voxel size, axial acquisition.
- moving: PD.2 follow-up exam 0.9375 x 0.9375 x 3 mm voxel size, axial acquisition.
- moving: T2.2 follow-up exam 0.9375 x 0.9375 x 3 mm voxel size, axial acquisition. -> same orientation as PD2, will have same transform applied
- moving:T1.2-GdDTPA follow-up exam0.9375 x 0.9375 x 5 mm voxel size, axial acquisition. -> undergoes 2 transforms: first to PD.2, then to PD.1
Registration Challenges
- we have multiple nested transforms: each exam is co-registered within itself, and then the exams are aligned to eachother
- potential pathology change can affect the registration
- anisotropic voxel size causes difficulty in rotational alignment
- clipped FOV and low tissue contrast of the post-contrast scan
Key Strategies
- we first register the post-contrast scans within each exam to the PD
- second we register the follow-up PD scan to the baseline PD
- we also move the T2 exam within the same Xform
- we then nest the first alignment within the second
- because of the contrast differences and anisotropic resolution we use Mutual Information as cost function for better robustness
Procedure
- Load & Center
- Load example dataset via OpenScene...
- Note that when you load the data as a scene, the image volumes have deliberately not been centered to demonstrate the effect of different image origins. Hence the result transforms provided will also not look right until the centering below is performed:
- Go to the Data module. You should see 6 images (e1_PD, e1_T2 etc.) and 3 solution transforms (Xform_...)
- Set background view to e1_PD and foreground to e2_PD. Toggle to see misalignment
- Center All images
- original images come with different origin settings, which make viewing difficult. We first re-center all images:
- Go to Volumes module, open Info tab
- From Active Volume menu, select each image in turn, then click the Center Image button
- Align Exam 1: FLAIR 1st pass: unmasked
- Open Registration / BRAINSFit module
- To set all parameters from presets, from the ParameterSet menu, select "Xf1_e1_FLAIR-T1_unmasked", else choose settings below:
- Fixed Image: e1_T1, moving image: e1_FLAIR
- Registration Phases: select "Initialize with CenterOfHeadAlign", Include Rigid, "IncludeScaleVersor3D" and Include Affine
- Output Settings: under SlicerLinear Transform, select "Create New Linear Transform, then select Rename" and rename it to Xf1_e1_FLAIR-T1
- Registration Parameters: increase the Number of Samples field to 200,000
- Leave all other settings at defaults & Click: Apply. Registration should complete within ~ 30 seconds
- Go back to the Data module: you should see the e1_FLAIR image moved under the newly created transform
- Select "E1_T1" as background and e1_FLAIR as new foreground, toggle to see alignment
- you will notice a small but distinct residual translational misalignment, apparent on axial and sagittal slices. This likely arises from dominant edges of the skull. We rerun a 2nd pass with masks
- Align Exam 1: FLAIR 2nd pass: masked
- we obtain a mask for the moving image (e1_FLAIR) by sending the mask for the fixed image (e1_ICC) through the inverse of the above transform
- From File menu, select Add Data & reload a second copy of the above transform Xf1_e1_FLAIR-T1 and the labelmap "e1_ICC.nrrd"
- rename Xform to " Xf1i_e1_T1-FLAIR and the mask to "e1_FLAIR_mask"
- go to the Transforms module, select the Xf1i_e1_T1-FLAIR transform and then click on the Invert button
- go to the Data module and drag the node for "e1_FLAIR_mask" inside the transform node Xf1i_e1_T1-FLAIR
- right click on the Xf1i_e1_T1-FLAIR node and select Harden Transform. We now have an approximate mask for the moving FLAIR image.
- Open Registration / BRAINSFit module
- To set all parameters from presets, from the ParameterSet menu, select "Xf1c_e1_FLAIR-T1_masked", else choose settings below:
- Fixed Image: e1_T1, moving image: e1_FLAIR
- Registration Phases: select Include Rigid, "IncludeScaleVersor3D" and Include Affine. Make sure no initialization phases are selected.
- Output Settings: under SlicerLinear Transform, select "Create New Linear Transform, then select Rename" and rename it to Xf1c_e1_FLAIR-T1_masked
- Registration Parameters: increase the Number of Samples field to 200,000
- Control of Mask Processing tab: check "ROI" box,
- Input Fixed Mask, select "e1_ICC",
- Input Moving Mask, select "e1_FLAIR_mask"
- Leave all other settings at defaults & Click: Apply. Registration should complete within ~ 40 seconds
- You should see the earlier residual misalignment mostly gone.
- Align Exam 1: T2
- The T2 image is not far in pose from the T1 and also does not have the same skull-contrast issue as the FLAIR, we register directly to the T1 w/o masking
- Open Registration / BRAINSFit module
- To set all parameters from presets, from the ParameterSet menu, select "Xf2_e1_T2-T1_unmasked", else choose settings below:
- Fixed Image: e1_T1, moving image: e1_T2
- Registration Phases: select Include Rigid, "IncludeScaleVersor3D" and Include Affine.
- Output Settings: under SlicerLinear Transform, select "Create New Linear Transform, then select Rename" and rename it to Xf2_e1_T2-T1
- Registration Parameters: increase the Number of Samples field to 200,000
- Leave all other settings at defaults & Click: Apply. Registration should complete within ~ 30 seconds
- Align Exam 2:
- repeat the above steps for exam 2, i.e. align e2_FLAIR and e2_T2 with e2_T1
- the above issue with the FLAIR-T1 residual does not arise for the follow-up exam, hence direct registration (unmasked) is possible
- Registration presets: BRAINSFit see Xf3_ and Xf4_ for FLAIR and T2, respectively
the MRML node tree shows the hierarchy of multiple transfoms.
- Align Exam 2-1:
- finally we align the T1 references of exam 1 and 2, i.e. we align e2_T1 with e1_T1:
- Registration presets: BRAINSFit see Xf5_
- parameters: defaults except increase sample rate to 200,000.
- here we have genuine masks for both images: Control of Mask Processing Tab:
- Control of Mask Processing tab: check "ROI" box,
- Input Fixed Mask, select "e1_ICC",
- Input Moving Mask, select "e2_ICC"
- Leave all other settings at defaults & Click: Apply. Registration should complete within ~ 20 seconds
- Combine Transforms:
- to align all images in a common space, we combine a hierarchy of transforms, i.e. the e2_FLAIR undergoes both the Xf3_e2_FLAIR-T1 and the Xf4_e2-e1: See image on the right for what the final hierarchy should look like.
- right-click on the nested transforms and select Harden Transforms from the pulldown menu. Rename the hardened transforms to indicate the direct alignment with e1_T1.
- Resample
- resample all images with their respective transforms, using the Filtering -> Resample Scalar/Vector/DWI Volume module.
- within the module, select "ws" as interpolation type
- Nonrigid extension
- there is some residual distortion in the alignment of e2_T1 to e1_T1. Optionally one can address this via another separate nonrigid BSpline registration. However combining the Affine with the BSpline is not straightforward (as of v.3.6.1), to do this follow this outline:
- resample e2_T1 according to Xf5 : e2_T1_Xf5
- Perform a BSpline only alignment : Xf6_e2-e1_BSpline that aligns e2_T1_Xf5 with e1_T1. Grid size 3-5.
- Save resulting Xform
- Open Xform file and other saved affine transforms in Text Editor
- Paste the affine portion at the bottom of the BSpline and save under new name.
Registration Results
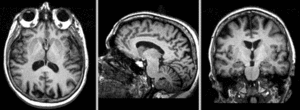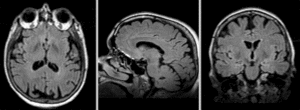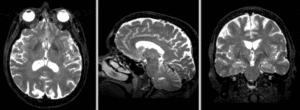 Exam 1 co-registered: FLAIR and T2 aligned with T1
Exam 1 co-registered: FLAIR and T2 aligned with T1
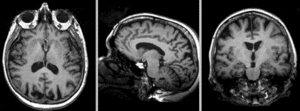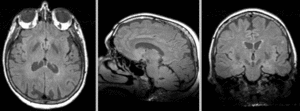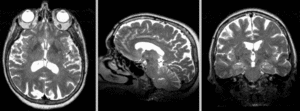 Exam 1 co-registered: FLAIR and T2 aligned with T1
Exam 1 co-registered: FLAIR and T2 aligned with T1
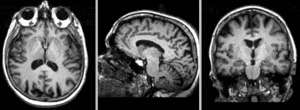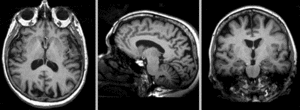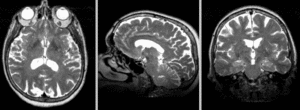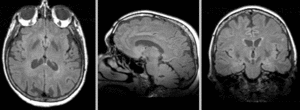 Exam 2 aligned to Exam 1 (showing FLAIR and T2 of exam 2 also)
Exam 2 aligned to Exam 1 (showing FLAIR and T2 of exam 2 also)