Difference between revisions of "Projects:RegistrationLibrary:RegLib C31"
From NAMIC Wiki
| Line 37: | Line 37: | ||
*'''Align Exam 2 to Exam 1: T1''' 1st pass: unmasked | *'''Align Exam 2 to Exam 1: T1''' 1st pass: unmasked | ||
#Open ''Registration / BRAINSFit'' module | #Open ''Registration / BRAINSFit'' module | ||
| − | + | ##Fixed Image: T1_e1, moving image: T1_e2 | |
| − | ##Fixed Image: | ||
##Registration Phases: select "Initialize with CenterOfHeadAlign", ''Include Rigid'', "IncludeScaleVersor3D" and ''Include Affine'' | ##Registration Phases: select "Initialize with CenterOfHeadAlign", ''Include Rigid'', "IncludeScaleVersor3D" and ''Include Affine'' | ||
| − | ##Output Settings: under SlicerLinear Transform, select "Create New Linear Transform'', then select ''Rename" and rename it to '' | + | ##Output Settings: under SlicerLinear Transform, select "Create New Linear Transform'', then select ''Rename" and rename it to ''Xf1_e2-e1_Affine'' |
##Registration Parameters: increase the ''Number of Samples'' field to 200,000 | ##Registration Parameters: increase the ''Number of Samples'' field to 200,000 | ||
##Leave all other settings at defaults & Click: ''Apply''. Registration should complete within ~ 30 seconds | ##Leave all other settings at defaults & Click: ''Apply''. Registration should complete within ~ 30 seconds | ||
| − | ##Go back to the ''Data'' module: you should see the | + | ##Go back to the ''Data'' module: you should see the T1_e2 image moved under the newly created transform |
| − | ##Select " | + | ##Select "T1_e1" as background and ''T2_e2'' as new foreground, toggle to see alignment |
| − | ## | + | ##due to the strong deformations an affine alignment alone does not achieve a perfect match. We can extend the DOF to non-rigid BSpline transforms, but must be aware that doing so might remove differences of interest: |
| − | *'''Align Exam | + | *'''Align Exam 2:''' 2nd pass: nonrigid |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
#Open ''Registration / BRAINSFit'' module | #Open ''Registration / BRAINSFit'' module | ||
| − | + | ##Fixed Image: T1_e1, moving image: T1_e2 | |
| − | ##Fixed Image: | + | ##From "Initialize with previously generated transform" & select the above affine ''Xf1_e2-e1_Affine'' |
| − | ## | + | ##Registration Phases: select only ''Include BSpline''. |
| − | ##Output Settings: under | + | ##Output Settings: under SlicerBSpline Transform, select "Create New BSpline Transform'', then select ''Rename" and rename it to ''Xf2_e2-e1_BSpl'' |
##Registration Parameters: increase the ''Number of Samples'' field to 200,000 | ##Registration Parameters: increase the ''Number of Samples'' field to 200,000 | ||
| − | + | ##Leave all other settings at defaults & Click: ''Apply''. Registration should complete within 3 minutes. | |
| − | + | ##You should see the earlier residual misalignment reduced, but strong distortions applied to the e2 image (see results below) | |
| − | |||
| − | ##Leave all other settings at defaults & Click: ''Apply''. Registration should complete within | ||
| − | ##You should see the earlier residual misalignment | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
Revision as of 16:10, 3 November 2010
Home < Projects:RegistrationLibrary:RegLib C31Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.1  Slicer Registration Library Case xx: TBI
Slicer Registration Library Case xx: TBI
Input
Modules
- Slicer 3.6.1 recommended modules: BrainsFit, Expert Automated Registration
Objective / Background
We seek to align two exams (acute baseline and follow-up) as well as all series within exams into a common space for direct evaluation of regional change.
Input Data
- inter-exam
- reference/fixed : T1_e1 baseline exam of acute TBI , 512 x 512 x 160 ; 0.5 x 0.5 x 1 mm voxel size, axial acquisition, RAS orientation.
- moving:T1_e2 follow-up exam of acute TBI , 512 x 512 x 160 ; 0.5 x 0.5 x 1 mm voxel size, axial acquisition, RAS orientation.
- intra-exam
Registration Challenges
- we have multiple nested transforms: each exam is co-registered within itself, and then the exams are aligned to eachother
- strong pathology (hemorrhage?) present at different amounts in the two exams
Key Strategies
- we first register the scans within each exam to the T1
- second we register the follow-up T1 scan to the baseline T1
- we then nest the first alignment within the second
Procedure
- Load & Center
- Load T1 MPRAGE datasets via Load Volume...
- volumes are note well centered, i.e. their physical origin defined in the image header differs; we therefore first center both images:
- Go to Volumes module, open Info tab
- From Active Volume menu, select each image in turn, then click the Center Image button
- note how Image origin changes for T1_e1 from 121,-97,-97 to 128,-128,-80 and for T1_e2 from 129,-106,-66 to 128,-128,-80
- now that both images have same center their initial misalignment can be seen by placing T1_e1 in the background and T1_e in the foreground and using the toggle-switch
- Align Exam 2 to Exam 1: T1 1st pass: unmasked
- Open Registration / BRAINSFit module
- Fixed Image: T1_e1, moving image: T1_e2
- Registration Phases: select "Initialize with CenterOfHeadAlign", Include Rigid, "IncludeScaleVersor3D" and Include Affine
- Output Settings: under SlicerLinear Transform, select "Create New Linear Transform, then select Rename" and rename it to Xf1_e2-e1_Affine
- Registration Parameters: increase the Number of Samples field to 200,000
- Leave all other settings at defaults & Click: Apply. Registration should complete within ~ 30 seconds
- Go back to the Data module: you should see the T1_e2 image moved under the newly created transform
- Select "T1_e1" as background and T2_e2 as new foreground, toggle to see alignment
- due to the strong deformations an affine alignment alone does not achieve a perfect match. We can extend the DOF to non-rigid BSpline transforms, but must be aware that doing so might remove differences of interest:
- Align Exam 2: 2nd pass: nonrigid
- Open Registration / BRAINSFit module
- Fixed Image: T1_e1, moving image: T1_e2
- From "Initialize with previously generated transform" & select the above affine Xf1_e2-e1_Affine
- Registration Phases: select only Include BSpline.
- Output Settings: under SlicerBSpline Transform, select "Create New BSpline Transform, then select Rename" and rename it to Xf2_e2-e1_BSpl
- Registration Parameters: increase the Number of Samples field to 200,000
- Leave all other settings at defaults & Click: Apply. Registration should complete within 3 minutes.
- You should see the earlier residual misalignment reduced, but strong distortions applied to the e2 image (see results below)
Registration Results
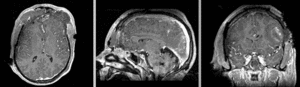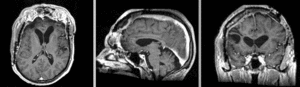
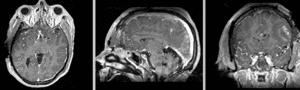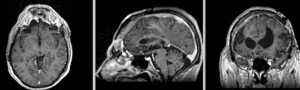 unregistered exam 1 & 2
unregistered exam 1 & 2
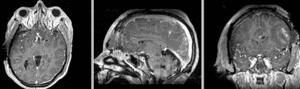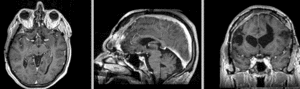 Exam 2 aligned to Exam 1 (affine only)
Exam 2 aligned to Exam 1 (affine only)
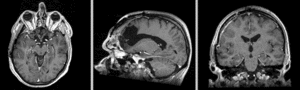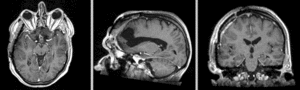
 Exam 2 aligned to Exam 1 (affine+BSpline)
Exam 2 aligned to Exam 1 (affine+BSpline)
 BSpline deformation only of Exam 2
BSpline deformation only of Exam 2