Difference between revisions of "Projects:RegistrationLibrary:RegLib C34"
From NAMIC Wiki
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (2 intermediate revisions by one other user not shown) | |||
| Line 16: | Line 16: | ||
=== Modules === | === Modules === | ||
| − | *'''Slicer 3.6.1 recommended modules:''' [ | + | *'''Slicer 3.6.1 recommended modules:''' [https://www.slicer.org/wiki/Modules:BRAINSFit '''BrainsFit'''], [https://www.slicer.org/wiki/Modules:FastMarchingSegmentation-Documentation-3.6 '''Fast Marching Segmentation'''], [https://www.slicer.org/wiki/Modules:HistogramMatching-Documentation-3.6 '''Histogram Matching'''], [https://www.slicer.org/wiki/Modules:ModelMaker-Documentation-3.6 '''Model Maker'''], |
===Objective / Background === | ===Objective / Background === | ||
| Line 33: | Line 33: | ||
===Input Data=== | ===Input Data=== | ||
| − | *reference/fixed: | + | *reference/fixed MRI: 0.08 x 0.08 x 0.09 mm, 256 x 340 x 128 |
| − | * | + | *moving MRI: 0.08 x 0.08 x 0.09 mm, 256 x 340 x 128 |
| − | |||
===Registration Challenges=== | ===Registration Challenges=== | ||
| Line 42: | Line 41: | ||
===Key Strategies=== | ===Key Strategies=== | ||
| − | *'''Slicer 3.6.1 recommended modules: [ | + | *'''Slicer 3.6.1 recommended modules: [https://www.slicer.org/wiki/Modules:BRAINSFit BrainsFit] |
=== Procedures === | === Procedures === | ||
| − | *'''Phase I: | + | *'''Phase I: Histogram Normalization and Datatype conversion''' |
| − | # | + | #The original images have intensities ranging from -0.0007 to 0.003, which causes trouble with many modules that round this to zero |
| − | # | + | #we first shift the intensity range to integer range [0,1000] and convert to unsigned short or something equivalent |
| − | : | + | #one way to accomplish this in slicer is to perform a histogram matching with a reference volume that has a usable dynamic range |
| − | *'''Phase II: | + | #Load a reference volume and then use the [https://www.slicer.org/wiki/Modules:HistogramMatching-Documentation-3.6 '''Histogram Matching'''] module to adjust the dynamic range |
| − | # | + | #use the [https://www.slicer.org/wiki/Modules:CastImage-Documentation-3.6 '''Cast module'''] to convert the datatype to unsigned short. |
| − | # | + | #save the new volumes |
| − | # | + | *'''Phase II: Brain Segmentation''' |
| − | *'''Phase | + | #to obtain a usable mask for both images we first need to segment the two brains |
| − | # | + | #go to the [https://www.slicer.org/wiki/Modules:FastMarchingSegmentation-Documentation-3.6 '''Fast Marching Segmentation'''] module |
| + | #select 2-3 fiducials within the brain (see example dataset .fcv files), set a target size of ~ 0.8 ml and click Apply. | ||
| + | #save the two labelmaps for the MD1 and MD2 segmentations. These will serve as masks for registration and a guides for initial alignment | ||
| + | *'''Phase III: Manual Initial Alignment''' | ||
| + | #if two datasets have large initial misalignment and also different image content and unusual intensities, this makes it difficult for an automated initializer to find the proper position. This increases the likelihood for the automated registration to go awry. To avoid this we adjust the main (translational) misalignment manually. You may skip this step if your data has closer initial alignment | ||
| + | #for complex geometries a 3D model can be very helpful in judging alignment. Both VolumeRendering and surface models are viable options. Since we already have two brain segmentations we use these to build 2 surface models: | ||
| + | # use the [https://www.slicer.org/wiki/Modules:ModelMaker-Documentation-3.6 '''Model Maker'''] module to build two models for MD1 and MD2 brain segmentation | ||
#Go to the ''Data'' module, right-click on the "Scene" node and select "Insert Transform Node" | #Go to the ''Data'' module, right-click on the "Scene" node and select "Insert Transform Node" | ||
| − | #select the new transform node and then rename it to " | + | #select the new transform node and then rename it to "Xf1_ManualInit", in the ''MRML Node Inspector" (press return after typing) |
| − | #place the | + | #in the ''Data'' module, move the MD2 volume (and model) inside the new transform node |
| + | #place the MD1 in the background and the MD2 image in the foreground | ||
#move the fade slider halfway to the middle so that you can see both volumes | #move the fade slider halfway to the middle so that you can see both volumes | ||
| − | #go to the ''Transforms'' module, move the translation sliders to | + | #go to the ''Transforms'' module, move the translation & rotation sliders to get the two images/modules into approximate alignment. Since this serves as initialization only, alignment need not be perfect. |
| − | |||
*'''Phase IV: Affine Registration''' | *'''Phase IV: Affine Registration''' | ||
#you may be able to skip this step and perform affine + nonrigid registration in one step, however the stepwise approach gives you more insight and control | #you may be able to skip this step and perform affine + nonrigid registration in one step, however the stepwise approach gives you more insight and control | ||
#Go to the ''BRAINSFit'' module | #Go to the ''BRAINSFit'' module | ||
## enter the following settings: | ## enter the following settings: | ||
| − | ##Fixed Image: | + | ##Fixed Image: MD1, Moving Image: MD2 |
| − | ##Registration phases: from ''Initialize with previously generated transform', select " | + | ##Registration phases: from ''Initialize with previously generated transform', select "Xf1_ManualInit" node created before. |
##Registration phases: check boxes for ''Include Rigid registration phase'', ''Include Scale Versor3D registration phase'' ''Include Affine registration phase''. To combine with Phase V below also check the ''Include BSpline phase'' box. | ##Registration phases: check boxes for ''Include Rigid registration phase'', ''Include Scale Versor3D registration phase'' ''Include Affine registration phase''. To combine with Phase V below also check the ''Include BSpline phase'' box. | ||
| − | ##Output: Slicer Linear transform: create new, rename to " | + | ##Output: Slicer Linear transform: create new, rename to "Xf2_Affine_masked" |
##Registration Parameters: increase ''Number Of Samples'' to 200,000 | ##Registration Parameters: increase ''Number Of Samples'' to 200,000 | ||
| − | ##Control Of Mask Processing Tab: check ''ROI'' box, ''Input Fixed Mask'' : | + | ##Control Of Mask Processing Tab: check ''ROI'' box, ''Input Fixed Mask'' : MD1_fmseg; ''Input Moving Mask'': MD2_fmseg |
##Leave all other settings at default | ##Leave all other settings at default | ||
| − | ##click apply | + | ##click apply |
| − | #go to ''Data'' module. You should see the " | + | #go to ''Data'' module. You should see the "MD2" volume moved inside the "Xf2.." transform node |
| − | |||
*'''Phase V: Nonrigid Registration''' | *'''Phase V: Nonrigid Registration''' | ||
#Go to the ''BRAINSFit'' module | #Go to the ''BRAINSFit'' module | ||
##enter the following settings: | ##enter the following settings: | ||
| − | ##Fixed Image: | + | ##Fixed Image: MD1, Moving Image: MD2 |
| − | ##Registration phases: from ''Initialize with previously generated transform', select " | + | ##Registration phases: from ''Initialize with previously generated transform', select "Xf2_..." node created before. |
##Registration phases: check boxes for ''Include BSpline registration phase'' | ##Registration phases: check boxes for ''Include BSpline registration phase'' | ||
| − | ##Output: Slicer BSpline transform: create new, rename to " | + | ##Output: Slicer BSpline transform: create new, rename to "Xf3_BSpline_masked" |
| − | ##Output Image Volume: create new, rename to " | + | ##Output Image Volume: create new, rename to "MD2_Xf3"; ''Pixel Type'': "short" |
| − | ##Registration Parameters: increase ''Number Of Samples'' to 200,000; ''Number of Grid Subdivisions'': | + | ##Registration Parameters: increase ''Number Of Samples'' to 200,000; ''Number of Grid Subdivisions'': 5,5,5 |
| − | ##Control Of Mask Processing Tab: check ''ROI'' box, ''Input Fixed Mask'' : | + | ##Control Of Mask Processing Tab: check ''ROI'' box, ''Input Fixed Mask'' : MD1_fmseg; ''Input Moving Mask'': MD2_fmseg |
##Leave all other settings at default | ##Leave all other settings at default | ||
| − | ##click apply | + | ##click apply |
| − | + | #Extend of nonrigid alignment will depend on the application. For better match the gridsize can be increased to 9x9x9 and higher, along with the number of sample points. Because larger grid sizes bear the risk of unstable and unfeasible deformations, an iterative approach with increasing DOF is recommended. Since the current version (3.6.1) of Slicer does not yet support concatenation of nonrigid transforms, such a stepwise approach implies multiple resampling of the moving volume and associated interpolation blurring. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
[[Image:RegLib_C34_AGif_Model.gif |200px|Surface model of original unregistered brains]] original, not registered<br> | [[Image:RegLib_C34_AGif_Model.gif |200px|Surface model of original unregistered brains]] original, not registered<br> | ||
Latest revision as of 18:07, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C34Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.1  Slicer Registration Library Case #34: Mouse Brain MRI
Slicer Registration Library Case #34: Mouse Brain MRI
Input
|
|
|
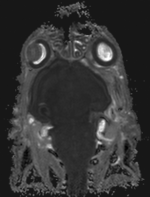
| fixed image 1/target MRI mouse 1 |
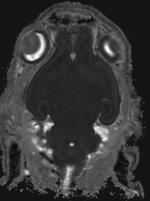
moving image MRI mouse 2 |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit, Fast Marching Segmentation, Histogram Matching, Model Maker,
Objective / Background
Aim is to register the brain of a 5 days old mouse (Post Natal Day 5, PND5) with an atlas.
Download
- DATA
- PRESETS:
- Documentation:
- See detailed pipeline with parameter settings below
Keywords
MRI, brain, mouse, masking, non-human, non-rigid,
Input Data
- reference/fixed MRI: 0.08 x 0.08 x 0.09 mm, 256 x 340 x 128
- moving MRI: 0.08 x 0.08 x 0.09 mm, 256 x 340 x 128
Registration Challenges
- The original images have very unusual dynamic range which can cause problems with modules that expect a minimal dynamic range in absolute terms
- target image content is of lower intensity than the surrounding tissue, making masking essential
Key Strategies
- Slicer 3.6.1 recommended modules: BrainsFit
Procedures
- Phase I: Histogram Normalization and Datatype conversion
- The original images have intensities ranging from -0.0007 to 0.003, which causes trouble with many modules that round this to zero
- we first shift the intensity range to integer range [0,1000] and convert to unsigned short or something equivalent
- one way to accomplish this in slicer is to perform a histogram matching with a reference volume that has a usable dynamic range
- Load a reference volume and then use the Histogram Matching module to adjust the dynamic range
- use the Cast module to convert the datatype to unsigned short.
- save the new volumes
- Phase II: Brain Segmentation
- to obtain a usable mask for both images we first need to segment the two brains
- go to the Fast Marching Segmentation module
- select 2-3 fiducials within the brain (see example dataset .fcv files), set a target size of ~ 0.8 ml and click Apply.
- save the two labelmaps for the MD1 and MD2 segmentations. These will serve as masks for registration and a guides for initial alignment
- Phase III: Manual Initial Alignment
- if two datasets have large initial misalignment and also different image content and unusual intensities, this makes it difficult for an automated initializer to find the proper position. This increases the likelihood for the automated registration to go awry. To avoid this we adjust the main (translational) misalignment manually. You may skip this step if your data has closer initial alignment
- for complex geometries a 3D model can be very helpful in judging alignment. Both VolumeRendering and surface models are viable options. Since we already have two brain segmentations we use these to build 2 surface models:
- use the Model Maker module to build two models for MD1 and MD2 brain segmentation
- Go to the Data module, right-click on the "Scene" node and select "Insert Transform Node"
- select the new transform node and then rename it to "Xf1_ManualInit", in the MRML Node Inspector" (press return after typing)
- in the Data module, move the MD2 volume (and model) inside the new transform node
- place the MD1 in the background and the MD2 image in the foreground
- move the fade slider halfway to the middle so that you can see both volumes
- go to the Transforms module, move the translation & rotation sliders to get the two images/modules into approximate alignment. Since this serves as initialization only, alignment need not be perfect.
- Phase IV: Affine Registration
- you may be able to skip this step and perform affine + nonrigid registration in one step, however the stepwise approach gives you more insight and control
- Go to the BRAINSFit module
- enter the following settings:
- Fixed Image: MD1, Moving Image: MD2
- Registration phases: from Initialize with previously generated transform', select "Xf1_ManualInit" node created before.
- Registration phases: check boxes for Include Rigid registration phase, Include Scale Versor3D registration phase Include Affine registration phase. To combine with Phase V below also check the Include BSpline phase box.
- Output: Slicer Linear transform: create new, rename to "Xf2_Affine_masked"
- Registration Parameters: increase Number Of Samples to 200,000
- Control Of Mask Processing Tab: check ROI box, Input Fixed Mask : MD1_fmseg; Input Moving Mask: MD2_fmseg
- Leave all other settings at default
- click apply
- go to Data module. You should see the "MD2" volume moved inside the "Xf2.." transform node
- Phase V: Nonrigid Registration
- Go to the BRAINSFit module
- enter the following settings:
- Fixed Image: MD1, Moving Image: MD2
- Registration phases: from Initialize with previously generated transform', select "Xf2_..." node created before.
- Registration phases: check boxes for Include BSpline registration phase
- Output: Slicer BSpline transform: create new, rename to "Xf3_BSpline_masked"
- Output Image Volume: create new, rename to "MD2_Xf3"; Pixel Type: "short"
- Registration Parameters: increase Number Of Samples to 200,000; Number of Grid Subdivisions: 5,5,5
- Control Of Mask Processing Tab: check ROI box, Input Fixed Mask : MD1_fmseg; Input Moving Mask: MD2_fmseg
- Leave all other settings at default
- click apply
- Extend of nonrigid alignment will depend on the application. For better match the gridsize can be increased to 9x9x9 and higher, along with the number of sample points. Because larger grid sizes bear the risk of unstable and unfeasible deformations, an iterative approach with increasing DOF is recommended. Since the current version (3.6.1) of Slicer does not yet support concatenation of nonrigid transforms, such a stepwise approach implies multiple resampling of the moving volume and associated interpolation blurring.
Registration Results
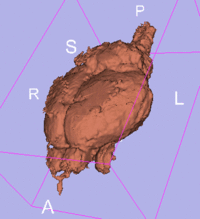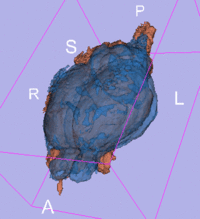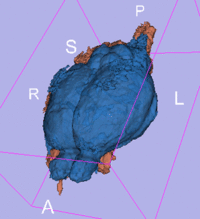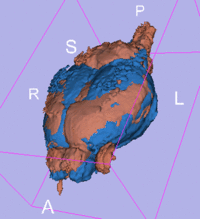 original, not registered
original, not registered
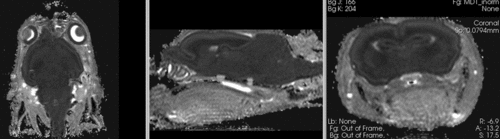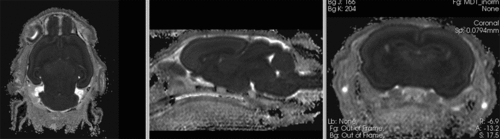 original, not registered
original, not registered
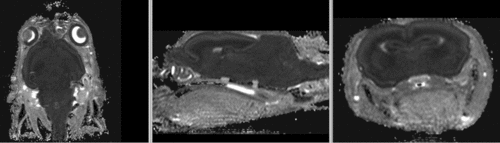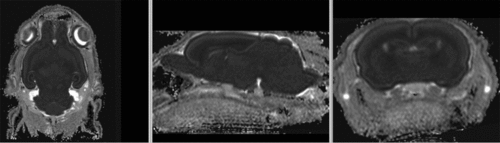 affine registered
affine registered
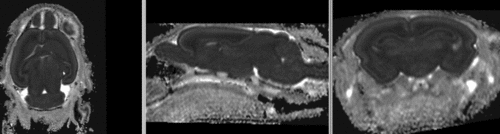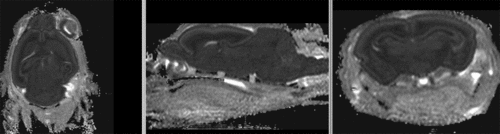 registered brains after 5x5x5 nonrigid BSpline alignment]
registered brains after 5x5x5 nonrigid BSpline alignment]