Difference between revisions of "Projects:RegistrationLibrary:RegLib C37"
From NAMIC Wiki
(Created page with 'Back to ARRA main page <br> Back to Registration main page <br> [[Projects:RegistrationDocumentation:UseCaseInv…') |
|||
| Line 7: | Line 7: | ||
{| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | ||
|[[Image:RegLib_C37_Thumb1.png|200px|lleft|this is the fixed reference image. All images are aligned into this space]] | |[[Image:RegLib_C37_Thumb1.png|200px|lleft|this is the fixed reference image. All images are aligned into this space]] | ||
| − | |[[Image: | + | |[[Image:RegArrow_Nonrigid.png|100px|lleft]] |
| − | |[[Image: | + | |[[Image:RegLib_C37_Thumb2.png|200px|lleft|this is the moving image. The transform is calculated by matching this to the reference image]] |
|- | |- | ||
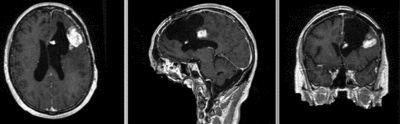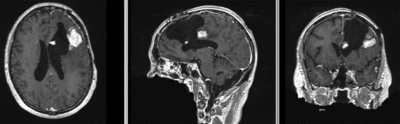
|fixed image/target | |fixed image/target | ||
| Line 31: | Line 31: | ||
=== Registration Results=== | === Registration Results=== | ||
{| style="color:#bbbbbb; background-color:#333333;" cellpadding="10" cellspacing="0" border="0" | {| style="color:#bbbbbb; background-color:#333333;" cellpadding="10" cellspacing="0" border="0" | ||
| − | |[[Image: | + | |[[Image:RegLib_C37_unreg.gif|400px|center|unregistered]] |
| − | |[[Image: | + | |[[Image:RegLib_C37_Xf1.gif|400px|center|rigid]] |
| − | |[[Image: | + | |[[Image:RegLib_C37_Xf2.gif|400px|center|affine]] |
| + | |[[Image:RegLib_C37_Xf3.gif|400px|center|BSpline (7x7x7) unmasked]] | ||
| + | |[[Image:RegLib_C37_Xf4.gif|400px|center|BSpline (7x7x7) masked]] | ||
|- | |- | ||
| − | | | + | |original unregistered |
| − | | | + | |rigid registered |
| − | | | + | |affine registered |
| + | |nonrigid registered (7x7x7 BSpline unmasked) | ||
| + | |nonrigid registered (7x7x7 BSpline masked) | ||
|} | |} | ||
===Download === | ===Download === | ||
| − | * | + | *Data: |
| − | **'''[[Media: | + | **'''[[Media:RegLib_C37_Data.zip|Image Data + solutions package <small> (Data, Solution, zip file qqMB) </small>]]''' |
| − | * | + | *Presets: |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | [[Projects:RegistrationDocumentation:ParameterPresetsTutorial|Link to User Guide: How to Load/Save Registration Parameter Presets]] | ||
| − | + | *Documentation | |
<!-- | <!-- | ||
comment | comment | ||
--> | --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 20:58, 3 March 2011
Home < Projects:RegistrationLibrary:RegLib C37Back to ARRA main page
Back to Registration main page
Back to Registration Case Library
Contents
v3.6.1  Slicer Registration Library Case #37:
Slicer Registration Library Case #37:
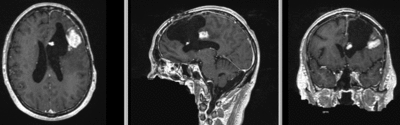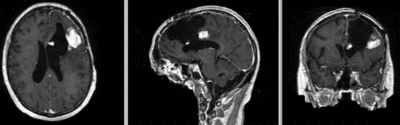
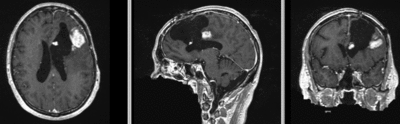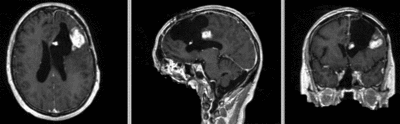
Intra-subject Brain MRI: T1 Tumor Growth / Resection Assessment
Input
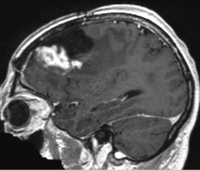
|
lleft | 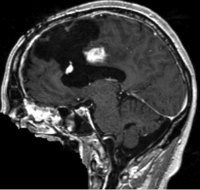
|
| fixed image/target | moving image |
Modules
Slicer 3.6.1 recommended modules:
Objective / Background
This is a typical case of change assessment.
Keywords
MRI, brain, head, intra-subject, T1, tumor, change assessment
Input Data
- reference/fixed : T1 SPGR , 1x1x1 mm voxel size, 256x256x176 ,sagittal
- moving: T1 SPGR , 1x1x1 mm voxel size, 256x256x176 ,sagittal
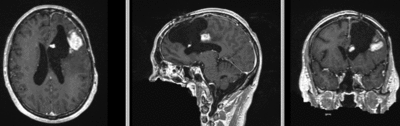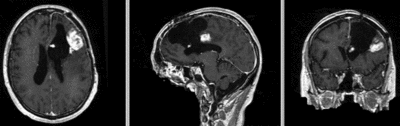
Registration Results
| original unregistered | rigid registered | affine registered | nonrigid registered (7x7x7 BSpline unmasked) | nonrigid registered (7x7x7 BSpline masked) |
Download
Link to User Guide: How to Load/Save Registration Parameter Presets
- Documentation