Difference between revisions of "Projects:RegistrationLibrary:RegLib C47"
From NAMIC Wiki
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (One intermediate revision by one other user not shown) | |||
| Line 16: | Line 16: | ||
=== Slicer4 Modules used === | === Slicer4 Modules used === | ||
| − | *[ | + | *[https://www.slicer.org/wiki/Documentation/4.1/Modules/BRAINSFit BrainsFit]''' |
===Objective / Background === | ===Objective / Background === | ||
| Line 22: | Line 22: | ||
===Download === | ===Download === | ||
| − | *[[Media:RegLib_C47_Data.zip|'''download input image data''' <small> ( | + | *[[Media:RegLib_C47_Data.zip|'''download RegLib_C47 input image data''' <small> (NRRD images, transforms, Slicer Scene File, zip file 52 MB) </small>]] |
=== Keywords === | === Keywords === | ||
| Line 40: | Line 40: | ||
*'''Phase I: MR-CTpre registration''' | *'''Phase I: MR-CTpre registration''' | ||
#Following the concept of manual registration, we create an initial transform that roughly aligns the MR to the pre-op CT. | #Following the concept of manual registration, we create an initial transform that roughly aligns the MR to the pre-op CT. | ||
| − | #open [ | + | #open [https://www.slicer.org/wiki/Documentation/4.1/Modules/BRAINSFit General Registraion (BRAINS)]''' module |
##''Input Images'' | ##''Input Images'' | ||
###''Fixed Image Volume'': CT_intraop | ###''Fixed Image Volume'': CT_intraop | ||
| Line 51: | Line 51: | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
##click: ''Apply'' | ##click: ''Apply'' | ||
| − | #switch to the [ | + | #switch to the [https://www.slicer.org/wiki/Documentation/4.1/Modules/Data Data module] |
##click on the "MRI_intra" node, and drag it onto the transform node "Xf1_MRI-CT_Affine" created above | ##click on the "MRI_intra" node, and drag it onto the transform node "Xf1_MRI-CT_Affine" created above | ||
#this should yield a rough alignment as shown in the result section below. We will use this to initialize a more refined nonrigid registration | #this should yield a rough alignment as shown in the result section below. We will use this to initialize a more refined nonrigid registration | ||
*'''Phase II: nonrigid registration''' | *'''Phase II: nonrigid registration''' | ||
| − | #open [ | + | #open [https://www.slicer.org/wiki/Documentation/4.1/Modules/BRAINSFit General Registraion (BRAINS)]'''module |
##''Input Images'': | ##''Input Images'': | ||
###''Fixed Image Volume'': CT_intraop | ###''Fixed Image Volume'': CT_intraop | ||
Latest revision as of 18:07, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C47Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
updated for v4.1 
Slicer Registration Library Case #47: Liver Tumor Cryoablation
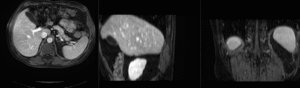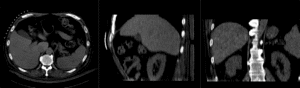
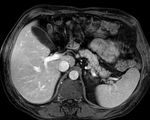
Input
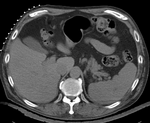
|
|
|
| fixed image/target | moving image |
Slicer4 Modules used
Objective / Background
We seek to align a pre-operative MRI with the intra-operative CT for surgical guidance.
Download
Keywords
MRI, CT, IGT, intra-operative, liver, cryoablation, change detection, non-rigid registration
Input Data
- reference/fixed : pr-op CT, 0.95 x 0.95 x 5 mm voxel size
- moving: intra-op MRI, 0.78 x 0.78 x 2.5 mm axial,
Discussion: Registration Challenges
- large differences in FOV
- strong differences in image contrast between MRI & CT
- we have strongly anisotropic voxel sizes with much less through-plane resolution
- there is a related example for pre-operative contrast CT to MRI in Library Case #12
Procedures
- Phase I: MR-CTpre registration
- Following the concept of manual registration, we create an initial transform that roughly aligns the MR to the pre-op CT.
- open General Registraion (BRAINS) module
- Input Images
- Fixed Image Volume: CT_intraop
- Moving Image Volume: MRI_preop
- Output Settings:
- Slicer BSpline Transform: none
- Slicer Linear Transform: (create new transform, rename to: "Xf1_MRI-CT_Affine")
- Output Image Volume: none
- Registration Phases: select/check Rigid , Rigid+Scale, Affine
- Leave all other settings at default
- click: Apply
- Input Images
- switch to the Data module
- click on the "MRI_intra" node, and drag it onto the transform node "Xf1_MRI-CT_Affine" created above
- this should yield a rough alignment as shown in the result section below. We will use this to initialize a more refined nonrigid registration
- Phase II: nonrigid registration
- open General Registraion (BRAINS)module
- Input Images:
- Fixed Image Volume: CT_intraop
- Moving Image Volume: MRI_preop
- Output Settings:
- Slicer BSpline Transform: (create new transform, rename to: "Xf2_MRI-CT_BSpline")
- Slicer Linear Transform: none
- Output Image Volume create new volume, rename to "MRI_Xf2"
- Initialization:
- Initialization transform: select "" created in phase 1 above
- Initialize Transform Mode: Off
- Registration Phases: select/check BSpline only
- Main Parameters:
- Number Of Samples: 200,000
- B-Spline Grid Size: 7,7,5
- Leave all other settings at default
- click: Apply
- Input Images:
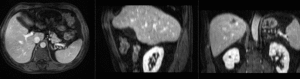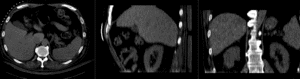
Registration Results (click to enlarge)
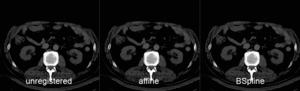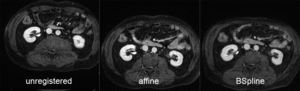
| unregistered MRI & CT | |
| after linear (affine) registration | |
| after nonrigid registration | |
| comparing kidney alignment at different registration stages |
Acknowledgments
Thanks to Dr.Stuart Silverman and Dr. Nobuhiko Hata for sharing this case.