Projects:RegistrationTBI
Back to Georgia Tech Algorithms
Multimodal Deformable Registration of Traumatic Brain Injury MR Volumes using Graphics Processing Units
Description
An estimated 1.7 million Americans sustain traumatic brain injuries (TBI's) every year. The large number of recent TBI cases in soldiers returning from military conflicts has highlighted the critical need for improvement of TBI care and treatment, and has drawn sustained attention to the need for improved methodologies of TBI neuroimaging data analysis. Neuroimaging of TBI is vital for surgical planning by providing important information for anatomic localization and surgical navigation, as well as for monitoring patient case evolution over time. Approximately 2 days after the acute injury, magnetic resonance imaging (MRI) becomes preferable to computed tomography (CT) for the purpose of lesion characterization, and the use of various MR sequences tailored to capture distinct aspects of TBI pathology provides clinicians with essential complementary information for the assessment of TBI-related anatomical insults and pathophysiology.
Image registration plays an essential role in a wide variety of TBI data analysis workflows. It aims to find a transformation between two image sets such that the transformed image becomes similar to the target image according to some chosen metric or criterion. Typically, a similarity measure is first established to quantify how `close` two image volumes are to each other. Next, the transformation that maximizes this similarity is typically computed through an optimization process which constrains the transformation to a predetermined class, such as rigid, affine or deformable. Numerous challenges associated with the task of TBI volume co-registration can exist if data acquisition is performed multimodally, and additional complexities can also arise due to the large degree of algorithmic robustness that may be required in order to properly address pathology-related deformations. Many conventional methods use the sum of squared differences of intensity values between two image sets as a similarity measure, which can perform poorly or even fail for TBI volume registration. Consequently, because the deformation of patient anatomy and soft tissues cannot typically be represented by rigid transforms, image registration often requires deformable image registration (DIR), i.e., the necessity of applying nonparametric infinite-dimensional transformations.
This paper proposes to replace the Mutual Information (MI) criterion for registration with the Bhattachayya distance [2] within a multimodal DIR framework [4]. The advantage of BD over MI is the superior behavior of the square root function compared to that of the logarithm at zero, which yields a more stable algorithm. This framework we describe takes into account the physical models of tissue motion to regularize the deformation fields and also involves free-form deformation. On the other hand, the DIR algorithm is computationally expensive when implemented on conventional central processing units, which can be detrimental particularly when three-dimensional (3D) volumes-rather than 2D images-need to be co-registered.
In clinical settings that involve acute TBI care, the amount of time required by the processing of neuroimaging data sets from patients in critical condition should be minimized. To meet this clinical requirement, we have implemented our algorithm on a graphics processing unit (GPU) platform.
Result
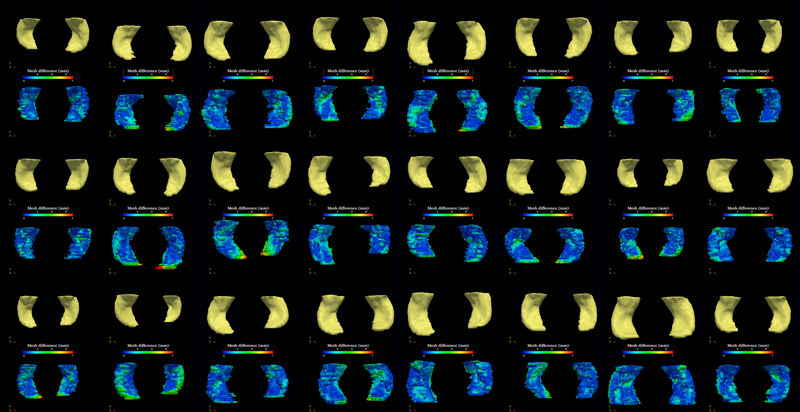
The segmentation method is applied on hippocampus. In the figure below, we show the segmentation results. In the first, third, and the fifth rows, the yellow colored shapes are the segmentation results output by the method. In the second, forth, and sixth rows, the colors on the shapes indicate the difference with the manual segmentation results: For each point on the shape (result of the segmentation algorithm), we compute the closest point on the manual segmented surface, and record the distance to that point. Such distances are encoded by the color shown in those rows.
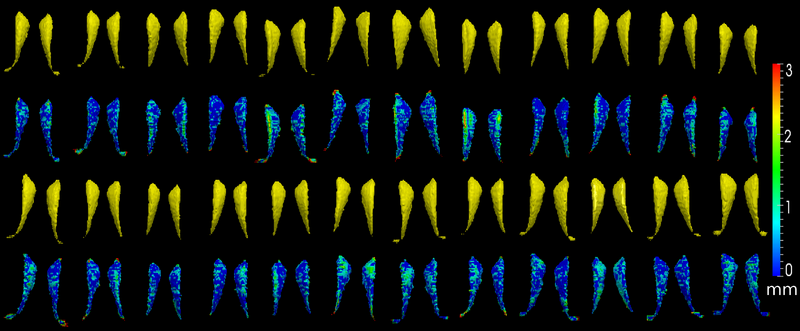
The method is also applied on caudate. Similarly to the hippocampus cases, in the figure below, we show the segmentation results. In the first and third rows, the yellow colored shapes are the segmentation results output by the method. In the second and forth rows, the colors on the shapes indicate the difference with the manual segmentation results: For each point on the shape (result of the segmentation algorithm), we compute the closest point on the manual segmented surface, and record the distance to that point. Such distances are encoded by the color shown in those rows.
Key Investigators
Georgia Tech: Yifei Lou and Patricio Vela Boston University: Allen Tannenbaum UCLA: Andrei Irimia, Micah C. Chambers, Jack Van Horn and Paul M. Vespa
References
1. Yifei Lou, Andrei Irimia, Patricio Vela, Allen Tannenbaum, Micah C. Chambers, Jack Van Horn and Paul M. Vespa. Multimodal Deformable Registration of Traumatic Brain Injury MR Volumes using Graphics Processing Units. In preparation. 2011
2. Yifei Lou and Allen Tannenbaum. Multimodal Deformable Image Registration via the Bhattacharyya Distance. Submitted to IEEE Trans. Image Process. 2011
3. Yifei Lou, Xun Jia, Xuejun Gu and Allen Tannenbaum. A GPU-based Implementation of Multimodal Deformable Image Registration Based on Mutual Information or Bhattacharyya Distance. Insight Journal, 2011. [online version][1]
4. E. D’Agostino, F. Maes, D. Vandermeulen, and P. Suetens. A viscous fluid model for multimodal non-rigid image registration using mutual information,” MICCAI, 2002, pp. 541–548