Projects:ShapeAnalysis
Back to NA-MIC Collaborations, MIT Algorithms,
Shape based Population Analysis
In contrast to shape-based segmentation that utilizes a statistical model of the shape variability in one population (typically based on the Principal Component Analysis), we are interested in identifying and characterizing differences between two sets of shape examples. We use the discriminative framework to characterize the differences in shape by training a classifier function and studying its sensitivity to small perturbations in the input data. An additional benefit of employing the classification approach is that the resulting classifier function can be used to label new examples into one of the two populations, e.g., for early detection in population screening or prediction in longitudinal studies. We estimate the expected accuracy of the classifier in a jackknife procedure. We have also adapted a non-parametric permutation test to the classification setting to estimate the statistical significance of the detected differences and the observed classification accuracy.
Description
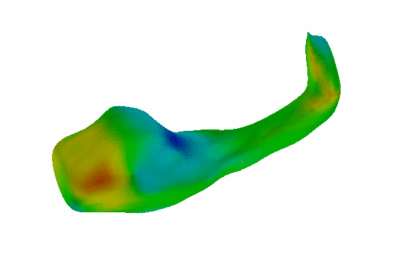
Study of Hippocampal Shape in Schizophrenia.
Recent Update
We have also started exploring alternative, surface-based descriptors for objects of spherical topology based on hierarchical decomposition of the surface functions into an over-complete basis. This work is still preliminary, but promises to improve our ability to detect and characterize subtle differences int he shape of anatomical structures due to diseases such as schizophrenia.
Structural Constellations for Population Analysis
We investigate a framework where global properties of structural constellations in medical images, i.e., the \emph{configuration and size} of multiple anatomical units, can be employed for population analysis of anatomical variability. The method takes advantage of the fact that cross-subject correspondence of certain structures is relatively well-established. This is in contrast with a majority of today's morphology studies that rely on local correspondence and/or employ a large number of features from each subject. Moreover, the representations we use can be interpreted in meaningful terms of global anatomy, allowing for the potential use of the analysis for exploring the pathology of neurodegenerative diseases like schizophrenia. In this paper, we demonstrate that with a small number of measurements per subject, one can achieve good separation between schizophrenics and matched controls. Our experiments indicate that the location of certain structures can capture discriminative characterization which may not be available through volumetric (size) measurements. For example, we find that the normalized sagittal position of the parahippocampal gyrus is significantly different between schizophrenics and controls. As an example, we also employ a linear Support Vector Machine to achieve up to 85% leave-one-out classification accuracy.
Software
- Stand alone code for training a classifier, jackknifing and permutation testing.
- Current plans include integration within the shape analysis pipeline, in collaboration with UNC (Martin Styner).
- We are currently also porting the software into ITK.
Publications
In Print