Projects:ShapeAnalysisFrameworkUsingSPHARMPDM
SPHARM-PDM
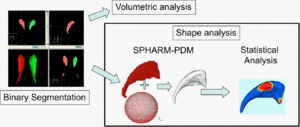
The UNC shape analysis is based on an analysis framework of objects with spherical topology, described by sampled spherical harmonics SPHARM-PDM. In summary, the input of the proposed shape analysis is a set of binary segmentation of a single brain structure, such as the hippocampus or caudate. These segmentations are first processed to fill any interior holes and a minimal smoothing operation. The processed binary segmentations are converted to surface meshes, and a spherical parametrization is computed for the surface meshes using a area-preserving, distortion minimizing spherical mapping. The SPHARM description is computed from the mesh and its spherical parametrization. Using the first order ellipsoid from the spherical harmonic coefficients, the spherical parametrizations are aligned to establish correspondence across all surfaces. The SPHARM description is then sampled into a triangulated surfaces (SPHARM-PDM) via icosahedron subdivision of the spherical parametrization. These SPHARM-PDM surfaces are all spatially aligned using rigid Procrustes alignment. Group differences between groups of surfaces are computed using the standard robust Hotelling T 2 two sample metric (see below). Statistical p-values, both raw and corrected for multiple comparisons, result in significance maps. Additional visualization of the group tests are provided via mean difference magnitude and vector maps, as well as maps of the group covariance information.
The implementation has reached a stable framework and has been disseminated to several collaborating labs within NAMIC (BWH, GeorgiaTech, Utah). The current development focuses on integrating the current command line tools into the Slicer (v3) via the Slicer execution model.