Difference between revisions of "Projects:TubularSurfaceSegmentation"
| Line 1: | Line 1: | ||
| − | + | Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm:GATech|Georgia Tech Algorithms]], [[Engineering:Kitware|Kitware Engineering]] | |
__NOTOC__ | __NOTOC__ | ||
= Tubular Surface Segmentation Framework = | = Tubular Surface Segmentation Framework = | ||
Revision as of 17:35, 17 July 2009
Home < Projects:TubularSurfaceSegmentationBack to NA-MIC Collaborations, Georgia Tech Algorithms, Kitware Engineering
Tubular Surface Segmentation Framework
This is a new model for tubular surfaces that transforms the problem of detecting a surface in 3D space, to detecting a curve in 4D space. This model affords computation efficiency and stability (by the use of the Sobolev norm) and we have successfully tested its application in segmenting the Cingulum Bundle from DW-MRI of the brain as well as vessel segmentation from CT cardiac data.
Description
We have proposed a new model for tubular surfaces that represents a tubular surface as a center-line with a radius function associated with every point of the center-line. This transforms the problem of detecting a surface in 3D space, to detecting a curve in 4D space. Besides allowing us to impose a "soft" tubular shape prior, this also leads to computational efficiency over conventional surface segmentation approaches. We have also developed the moving end points implementation of this framework wherein the required input is only a few points in the interior of the structure of interest. This yields the additional advantage that the framework simulatenously returns both the 3D segmentation and the 3D skeleton of the structure eliminating the need for apriori knowledge of end points, and an expensive skeletonization step. The framework is applicable to different tubular anatomical structures in the body. We have so far applied it successfully to the Cingulum Bundle, and blood vessels.
Some Results
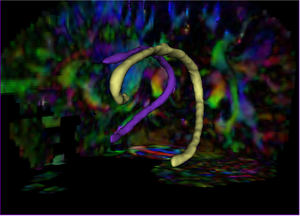 3D visualization of CB segmentation
3D visualization of CB segmentation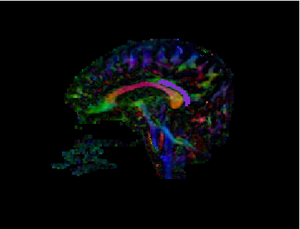 Slice-wise view of CB Segmentation result
Slice-wise view of CB Segmentation result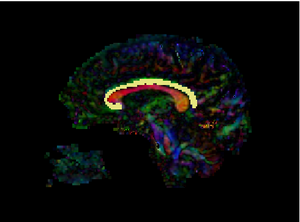 Slice-wise view of CB Segmentation result
Slice-wise view of CB Segmentation result
Project Status
- Algorithm successfully implemented and tested in MATLAB for Cingulum Bundle (as well as vessel segmentation).
Key Investigators
- Georgia Tech Algorithms: Vandana Mohan, Allen Tannenbaum
- Kitware Engineering: Luis Ibanez
Publications
- V. Mohan, G. Sundaramoorthi, J. Melonakos, M. Niethammer, M. Kubicki, and A. Tannenbaum. Tubular Surface Evolution for Segmentation of Tubular Structures with Applications to the Cingulum Bundle From DW-MRI. In Proceedings of MICCAI Workshop on Mathematical Foundations of Computational Anatomy, 2008.
- V. Mohan, G. Sundaramoorthi and A. Tannenbaum. Tubular Surface Segmentation for identifying anatomical structures from medical imagery. (In submission)