Difference between revisions of "Projects:TumorModeling"
(Created page with 'Back to NA-MIC Collaborations, MIT Algorithms __NOTOC__ = Modeling tumor growth in patients with glio…') |
|||
| Line 5: | Line 5: | ||
This opens two directions: First, it aims at making complex information from longitudinal multimodal data set accessible for diagnostic radiology through physiological models. This will allow to estimate features such as degree of infiltration, speed of growth or mass effect in a quantitative fashion; for therapy it will allow to identify regions at risk for progression. Second, it aims at providing the means to test different makroscopic tumor models from theoretical biology on real clinical data. | This opens two directions: First, it aims at making complex information from longitudinal multimodal data set accessible for diagnostic radiology through physiological models. This will allow to estimate features such as degree of infiltration, speed of growth or mass effect in a quantitative fashion; for therapy it will allow to identify regions at risk for progression. Second, it aims at providing the means to test different makroscopic tumor models from theoretical biology on real clinical data. | ||
| − | |||
| − | |||
| − | |||
| − | |||
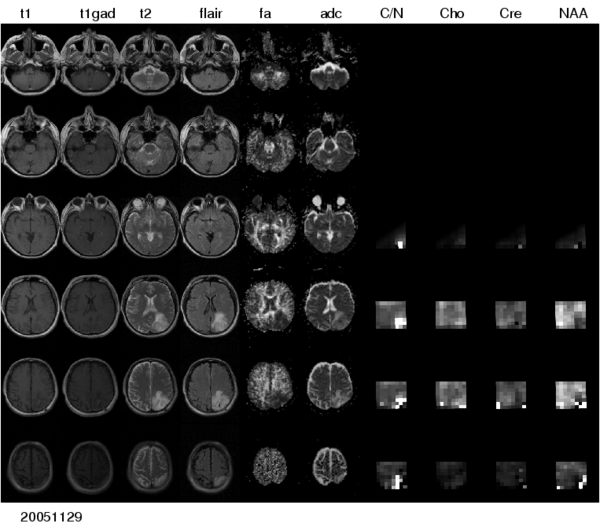
For realizing these aims, the project comprises a number of ascpectes -- automated segmentation of tumors in large multimodal image data sets, making information of different MR image modalities accessible for the tumor model, with a focus on the processing of magnetic resonance spectroscopic images (MRSI), and the development of methods for the image-based estimation of parameters in reaction-diffusion type models of tumor growth. | For realizing these aims, the project comprises a number of ascpectes -- automated segmentation of tumors in large multimodal image data sets, making information of different MR image modalities accessible for the tumor model, with a focus on the processing of magnetic resonance spectroscopic images (MRSI), and the development of methods for the image-based estimation of parameters in reaction-diffusion type models of tumor growth. | ||
| − | + | [[Image:Multimodal_glioma.png|thumb|center|600px| Figure 1: Multi-modal image data - different modalities available for a single timepoint.]] | |
| − | |||
| − | |||
| − | Image: | ||
| − | |||
| + | == Segmenting tumors in large multimodal data sets == | ||
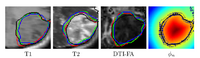
| + | [[Image:Tumor_segmentation_lesion_atlas.png|thumb|center|200px| Figure 1: Tumor segmentation]] | ||
To segment all MR image volumes available for a patient we developed an approach for learning patient-specific lesion atlases with limited user interaction. Figure 1 shows the manual segmentations of the tumor from different raters (red, green, blue) and the automatic segmentation using a patient-specific lesion atlas (black) in T1-MRI, T1-MRI and the fractional anisotropy map from DTI. The right image shows the lesion atlas. | To segment all MR image volumes available for a patient we developed an approach for learning patient-specific lesion atlases with limited user interaction. Figure 1 shows the manual segmentations of the tumor from different raters (red, green, blue) and the automatic segmentation using a patient-specific lesion atlas (black) in T1-MRI, T1-MRI and the fractional anisotropy map from DTI. The right image shows the lesion atlas. | ||
| − | = Processing magnetic resonance spectroscopic images = | + | == Processing magnetic resonance spectroscopic images == |
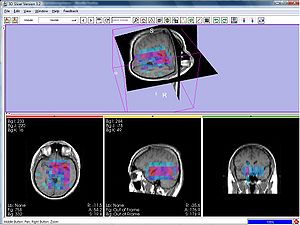
| − | + | [[Image:Mrsi parametric-map-NAA 3d.jpg|thumb|center|300px|Color-coded MRSI in the localization and grading of brain tumors]] | |
| − | Image:Mrsi | ||
| − | |||
| − | |||
| − | |||
| − | |||
To make the metabolic information of magnetic resonance spectroscopic images available for modeling the evolution of glioma growth we are implementing an [[http://wiki.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_MRSI-Module| MRSI processing module]] for Slicer. | To make the metabolic information of magnetic resonance spectroscopic images available for modeling the evolution of glioma growth we are implementing an [[http://wiki.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_MRSI-Module| MRSI processing module]] for Slicer. | ||
Revision as of 19:52, 10 September 2009
Home < Projects:TumorModelingBack to NA-MIC Collaborations, MIT Algorithms
Modeling tumor growth in patients with glioma
we are interested in developing computational methods for the assimilation of magnetic resonance image data into physiological models of glioma -- the most frequent primary brain tumor -- for a patient-adaptive modeling of tumor growth.
This opens two directions: First, it aims at making complex information from longitudinal multimodal data set accessible for diagnostic radiology through physiological models. This will allow to estimate features such as degree of infiltration, speed of growth or mass effect in a quantitative fashion; for therapy it will allow to identify regions at risk for progression. Second, it aims at providing the means to test different makroscopic tumor models from theoretical biology on real clinical data.
For realizing these aims, the project comprises a number of ascpectes -- automated segmentation of tumors in large multimodal image data sets, making information of different MR image modalities accessible for the tumor model, with a focus on the processing of magnetic resonance spectroscopic images (MRSI), and the development of methods for the image-based estimation of parameters in reaction-diffusion type models of tumor growth.
Segmenting tumors in large multimodal data sets
To segment all MR image volumes available for a patient we developed an approach for learning patient-specific lesion atlases with limited user interaction. Figure 1 shows the manual segmentations of the tumor from different raters (red, green, blue) and the automatic segmentation using a patient-specific lesion atlas (black) in T1-MRI, T1-MRI and the fractional anisotropy map from DTI. The right image shows the lesion atlas.
Processing magnetic resonance spectroscopic images
To make the metabolic information of magnetic resonance spectroscopic images available for modeling the evolution of glioma growth we are implementing an [MRSI processing module] for Slicer.
Key Investigators
- MIT: Bjoern Menze, Polina Golland
- INRIA Sophia-Antipolis, France: Ezequiel Geremia, Olivier Clatz, Nicholas Ayache
- DKFZ Heidelberg, Germany: Bram Stieltjes, Marc-Andre Weber