Difference between revisions of "Projects:UtahAtlasSegmentation"
Hjmjohnson (talk | contribs) |
Hjmjohnson (talk | contribs) |
||
| Line 57: | Line 57: | ||
*Utah Algorithms: Marcel Prastawa, Guido Gerig | *Utah Algorithms: Marcel Prastawa, Guido Gerig | ||
*UNC Algorithms: Martin Styner | *UNC Algorithms: Martin Styner | ||
| − | *External research collaboration: Hans Johnson, MIND Institute | + | *External research collaboration: Hans Johnson, The University of Iowa |
| + | *External research collaboration: Jeremy Bockholt, MIND Institute | ||
Revision as of 16:40, 11 May 2010
Home < Projects:UtahAtlasSegmentationBack to Utah 2 Algorithms
Atlas Based Brain Segmentation
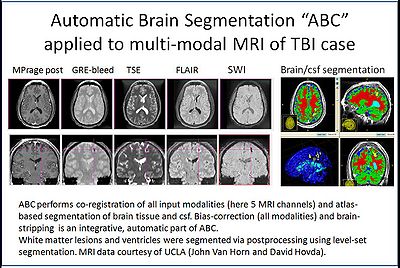
Automatic segmentation can be performed reliably using priors from brain atlases and an image generative model. We have developed a tool that provides an automatic segmentation pipeline in a modular framework. The processing pipeline is composed tasks such as filtering the input images, registering the multimodal input images and the brain atlas to a common space, followed by iterative steps which interleave segmentation, inhomogeneity correction, and atlas warping.
Our tool generates bias corrected images, fuzzy classification maps, and discrete segmentation labels. The tool has been used to automatically segment thousands of adult and toddler images from the University of North Carolina (UNC), and is also being used as a skull stripping mechanism for DTI processing at UNC and Utah. An example of the output of the tool is shown below.
The tool has been integrated into Slicer as an extension, and it can also be executed as a stand-alone application. Both versions are available for download through NITRC: http://www.nitrc.org/projects/abc .
External use and modifications of ABC
A predicessor of ABC has been distributed to a large number of other research groups associated with ongoing image analysis projects at UNC Chapel Hill and the University of Utah, and to research groups linked to these investigators. High profile projects include the Silvio Conte Center (http://www.psychiatry.unc.edu/conte/, PI John H. Gilmore, UNC) with processing of over 1000 infant MRI, and the Autism Centers of Excellence Project IBIS (http://www.ibis-network.org/, PI Joseph Piven, UNC) where over 650 infant brain MRI will be processed with this tool. To segment infant brains with age range 6months to 2 years as part of this longitudinal study, new age-specific atlases have been developed to be used as spatial priors for ABC.
The ABC method has also been modified and extensively used by the University of Iowa, Hans Johnson as part of a large, multi-center Huntington Disease study. Experience with a couple of hundred datasets at IOWA demonstrated the excellent robustness and reliability of the methodology.
The probability maps for the atlas were created from a set of 729 3T multi-modal image data sets (IOWA, Hans Johnson). Improvements made were to include a measure of Venus Blood as a part of the model, and to add explicit tissue regions outside the head for tissue types that are not part of the brain. This atlas definitions are available from svn co https://www.nitrc.org/svn/brains/BRAINS/trunk/BRAINSTools/BRAINSABC/Atlas_20100510
Under guidance of Hans Johnson, a branch of the ABC NITRC code has been modified to meet the needs of the IOWA group. This includes integration of a new Bspline-based registration from BRAINSFit, integration of BRAINSROIAuto, and to make it work with data with arbitrary orientation, and modifications of the interface (svn to https://www.nitrc.org/svn/brains/BRAINS/trunk/BRAINSTools/BRAINSABC).
ABC Bias Correction Module
To meet still strong demands for a stand-alone MRI bias correction method with FLASH images, part of the ABC code has been made available as a separate module and distributed via NITRC (http://www.nitrc.org/projects/probbiascor) by the MIND Institute group lead by Mark Skully and Jeremy Bockholt.
- Marcel Prastawa, John H. Gilmore, Weili Lin, and Guido Gerig. Automatic Segmentation of MR Images of the Developing Newborn Brain. Medical Image Analysis (MedIA). Vol 9, Issue 5, October 2005, Pages 457-466.
- Marcel Prastawa, Elizabeth Bullitt, Sean Ho, and Guido Gerig. A Brain Tumor Segmentation Framework Based on Outlier Detection . Medical Image Analysis (MedIA). Vol 8, Issue 3, September 2004, Pages 275-283.
- Marcel Prastawa, John Gilmore, Weili Lin, and Guido Gerig. Automatic Segmentation of Neonatal Brain MRI. Medical Image Computing and Computer Assisted Intervention (MICCAI) 2004. Lecture Notes in Computer Science (LNCS) 3216, Pages 10-17.
- Marcel Prastawa, Elizabeth Bullitt, Nathan Moon, Koen van Leemput, and Guido Gerig. Automatic Brain Tumor Segmentation by Subject Specific Modification of Atlas Priors. Academic Radiology. Vol 10, Issue 12, December 2003, Pages 1341-1348.
- Marcel Prastawa, Elizabeth Bullitt, Sean Ho, and Guido Gerig. Robust Estimation for Brain Tumor Segmentation. Medical Image Computing and Computer Assisted Intervention (MICCAI) 2003. Lecture Notes in Computer Science (LNCS) 2879, Pages 530-537.
Key Investigators
- Utah Algorithms: Marcel Prastawa, Guido Gerig
- UNC Algorithms: Martin Styner
- External research collaboration: Hans Johnson, The University of Iowa
- External research collaboration: Jeremy Bockholt, MIND Institute