Difference between revisions of "Slicer3:DTMRI"
From NAMIC Wiki
| Line 1: | Line 1: | ||
| − | = | + | = Goals = |
| − | Development of the infrastructure for DT-MRI processing and visualization | + | * Development of the core infrastructure for DT-MRI processing and visualization. |
| + | * Development of the core infrastructure for fiber tracks processing and visualization. | ||
| + | * Integration of new and existing methods and algorithms for DT-MRI processing using the provided infrastructure. | ||
| + | * Porting of the current DT-MRI capabilities existing in Slicer 2.x | ||
| − | = | + | = Core infrastructure for DT-MRI processing and visualization, fiber processing and visualization = |
| + | == DTI Data Model == | ||
| + | MRML nodes for different data representations involved in DTI analysis: | ||
| + | * Diffusion Weighted Images: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLDiffusionWeightedVolumeNode.h?view=log vtkMRMLDiffusionWeightedVolumeNode]. | ||
| + | * Diffusion Tensor Images: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLDiffusionTensorVolumeNode.h?view=log vtkMRMLDiffusionTensorVolumeNode]. | ||
| + | * Fiber Bundles: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLFiberBundleNode.h?view=log vtkMRMLFiberBundleNode]. | ||
| − | + | == Visualization/Display == | |
| + | MRML nodes for definition for DTI data visualization: | ||
| + | * Diffusion Weighted Images: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLDiffusionWeightedVolumeDisplayNode.h?view=log vtkMRMLDiffusionWeightedVolumeDisplayNode]. | ||
| + | * Diffusion Tensor Images: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLDiffusionTensorVolumeDisplayNode.h?view=log vtkMRMLDiffusionTensorVolumeDisplayNode], and [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLDiffusionTensorDisplayPropertiesNode.h?view=log vtkMRMLDiffusionTensorDisplayPropertiesNode]. | ||
| + | * Fiber Bundles: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLFiberBundleDisplayNode.h?view=log vtkMRMLFiberBundleDisplayNode], [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLFiberBundleLineDisplayNode.h?view=log vtkMRMLFiberBundleLineDisplayNode], [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLFiberBundleTubeDisplayNode.h?view=log vtkMRMLFiberBundleTubeDisplayNode], and [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLFiberBundleGlyphDisplayNode.h?view=log vtkMRMLFiberBundleGlyphDisplayNode]. | ||
| − | + | ==Storage and I/O == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
* DWI and DTI I/O: NRRD is the format supported by Slicer 3 for storing DWI and DTI images. | * DWI and DTI I/O: NRRD is the format supported by Slicer 3 for storing DWI and DTI images. | ||
** NNRD reader/writer: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem/vtkNRRDReader.h?view=log vtkNRRDReader] and [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem/vtkNRRDWriter.h?view=log vtkNRRDWriter]. | ** NNRD reader/writer: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem/vtkNRRDReader.h?view=log vtkNRRDReader] and [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem/vtkNRRDWriter.h?view=log vtkNRRDWriter]. | ||
| Line 25: | Line 25: | ||
* Fiber I/O: vtkPolyData has been the format adopted for the description of fibers. | * Fiber I/O: vtkPolyData has been the format adopted for the description of fibers. | ||
** Storage node: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLFiberBundleStorageNode.h?view=log vtkMRMLFiberBundleStorageNode] | ** Storage node: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/MRML/vtkMRMLFiberBundleStorageNode.h?view=log vtkMRMLFiberBundleStorageNode] | ||
| − | |||
| − | + | == Displaying Logic == | |
| − | * | + | * Visualization pipelines for DTI, DWI volumes, and fiber bundles are incorporated into the corresponding display nodes. |
| − | * | + | * DWI volumes are displayed as separate components. |
| + | * DTI volumes are displayed computed scalar properties (such as FA, Linear Measure, etc.) | ||
| + | * Fiber bundles are displayed as lines, tubes, and glyphs with their own properties and colors. | ||
| − | + | == Diffusion Processing Toolbox == | |
| − | + | [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem vtkTeem-library] provides tools for: | |
| − | + | * Tensor estimation | |
| − | + | * Computation of scalar measurements from tensor fields | |
| − | + | *Fast rendering of tensor fields using glyphs: line, box, ellipsoid. | |
| − | + | *Fiber Tracking using integration techniques | |
| + | * Multiple ROI seeding and logic interconnections between ROIs | ||
| + | |||
| + | = DT-MRI processing and visualization Modules = | ||
| + | |||
| + | == Tensor Estimation from DWI == | ||
| + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/DiffusionTensorEstimation.xml?view=log DiffusionTensorEstimation]. | ||
Teem currently provides a clean interface to do this estimation in a voxel by voxel fashion. Collaboration with Gordon Kindlmann for a vtk filter implementation that encapsulates the estimation process ([http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem/vtkTeemEstimateDiffusionTensor.h?view=log vtkTeemEstimateDiffusionTensor]). | Teem currently provides a clean interface to do this estimation in a voxel by voxel fashion. Collaboration with Gordon Kindlmann for a vtk filter implementation that encapsulates the estimation process ([http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem/vtkTeemEstimateDiffusionTensor.h?view=log vtkTeemEstimateDiffusionTensor]). | ||
| − | == | + | == Diffusion Tensor Scalar Measurements == |
| − | * | + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/DiffusionTensorMathematics.xml?view=log DiffusionTensorMathematics]. |
| − | * | + | * Implemented in vtkTeem library [http://www.na-mic.org/ViewVC/index.cgi/trunk/Libs/vtkTeem/vtkDiffusionTensorMathematics.h?view=log vtkDiffusionTensorMathematics]. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | == Rician LMMSE Filter == |
| + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/DiffusionApplications/dwiNoiseFilter/dwiNoiseFilter.xml?view=log dwiNoiseFilter]. | ||
| − | * | + | *Filters a set of diffusion weighted images in the mean squared error sense using a Rician noise model. The noise parameter is automatically estimated. |
| + | *Contributed by Santiago Aja Fernandez and Marc Niethammer | ||
| − | + | == Tractography Display/Load/Save Module== | |
| + | * Load and Saves fiber tracks as vtkPolyData files (.vtp) | ||
| + | * Displays fiber tracks as lines, tubes, glyphs. Allows to control line/tube/glyph display properties and colors | ||
| + | * Built-in Slicer3 module. | ||
| − | * | + | == Tractography ROI Seeding Module == |
| − | * | + | * Creates fibers passing through a specified ROI |
| − | * | + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Tractography/Seeding/Seeding.xml?view=log ROI Seeding]. |
| − | * | + | |
| + | == Tractography Fiducial Seeding Module == | ||
| + | * Creates fibers passing through the specified fiducial points interactively | ||
| + | * Built-in Slicer3 module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Tractography/FiducilaSeeding/vtkSlicerTractographyFiducialSeedingGUI.cxx?view=log vtkSlicerTractographyFiducialSeedingGUI.cxx]. | ||
| − | |||
| − | + | == Tractography ROI Select Module == | |
| + | * Select tracts passing or not passing through ROIs | ||
| + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Tractography/Editor/ROISelect.xml?view=log ROI Select]. | ||
| − | == | + | == Stochastic Tractography Filter == |
| + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/DiffusionApplications/StochasticTractographyFilter/StochasticTractographyFilter.xml?view=log StochasticTractographyFilter]. | ||
| + | * Generates a map of connectivity probabilities from a DWI volume.). | ||
| + | * Contributed by Tri Ngo (tringo@gmail.com) | ||
| − | + | == ROI Tract Filter == | |
| − | * | + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/DiffusionApplications/StochasticTractographyFilter/ROITractFilter.xml?view=log ROITractFilter]. |
| − | + | * Creates a new tract container containing only tracts which pass through the selected ROI's. | |
| − | ** | + | * Contributed by Tri Ngo (tringo@gmail.com) |
| − | + | == Generate Connectivity Map == | |
| − | * | + | * CLI Module: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/DiffusionApplications/StochasticTractographyFilter/GenerateConnectivityMap.xml?view=log GenerateConnectivityMap]. |
| − | + | * Generates a volume where the value of each voxel is the number of fibers which pass through that voxel divided by the total number of sampled fibers. This value can been interpreted as the probability that a particular voxel is connected to the seed ROI by a fiber tract. | |
| − | + | * Contributed by Tri Ngo (tringo@gmail.com) | |
| − | |||
| − | |||
| − | * | ||
| − | * | ||
| − | |||
| − | |||
| − | + | : | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | = | + | = Future development plans = |
| + | * Fiber editing: enviroment for manually editing individual fibers/bundles, reassigning of fibers to bundles. | ||
| + | * Teem based tractography (Core 1 & 2). | ||
| + | * Fiber Bundle Clustering (Core 1). | ||
| + | * Render glyphs in the 2D slice windows. | ||
| + | * Statistics along fiber tracts (Core 1). | ||
* Quantitative measurement | * Quantitative measurement | ||
** Tract-based | ** Tract-based | ||
| Line 97: | Line 103: | ||
* fMRI seeding | * fMRI seeding | ||
* Surgical planning | * Surgical planning | ||
| − | * | + | * DT-MRI segmentation/atlas creation: enviroment for segmentation of DT-MRI fields |
| − | + | * DT-MRI registration: enviroment for registration of DT-MRI fields (possibly via DWI registration -- work done at GE and presented in MICCAI '06). | |
| + | * Tensor estimation using different methods, namely: | ||
| + | ** Least Squares | ||
| + | ** Weighted Least Squares | ||
| + | ** Non-linear methods | ||
| + | ** Maximum Likelihood approach | ||
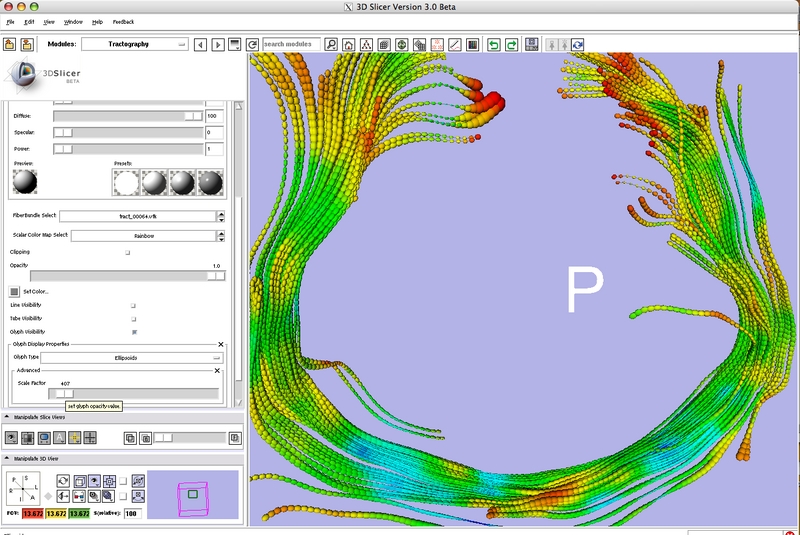
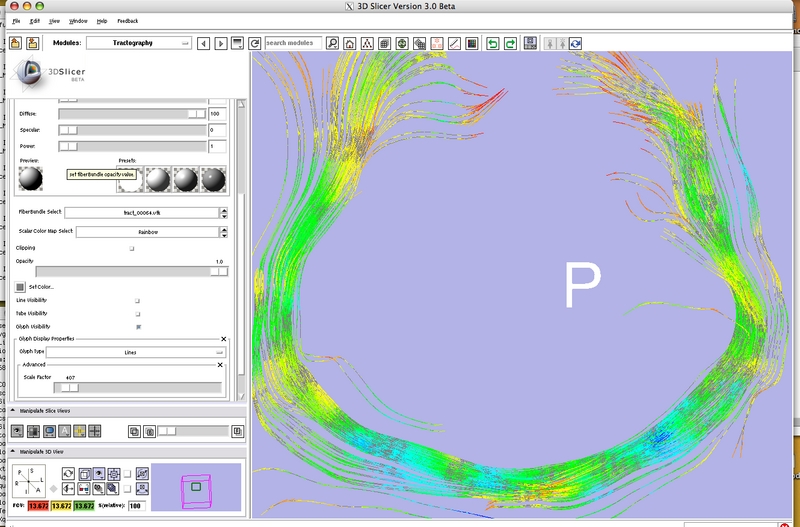
= Development Screenshots = | = Development Screenshots = | ||
Revision as of 21:50, 2 January 2008
Home < Slicer3:DTMRIContents
- 1 Goals
- 2 Core infrastructure for DT-MRI processing and visualization, fiber processing and visualization
- 3 DT-MRI processing and visualization Modules
- 3.1 Tensor Estimation from DWI
- 3.2 Diffusion Tensor Scalar Measurements
- 3.3 Rician LMMSE Filter
- 3.4 Tractography Display/Load/Save Module
- 3.5 Tractography ROI Seeding Module
- 3.6 Tractography Fiducial Seeding Module
- 3.7 Tractography ROI Select Module
- 3.8 Stochastic Tractography Filter
- 3.9 ROI Tract Filter
- 3.10 Generate Connectivity Map
- 4 Future development plans
- 5 Development Screenshots
- 6 Notes on general diffusion framework (ODF/2 tensor) support
Goals
- Development of the core infrastructure for DT-MRI processing and visualization.
- Development of the core infrastructure for fiber tracks processing and visualization.
- Integration of new and existing methods and algorithms for DT-MRI processing using the provided infrastructure.
- Porting of the current DT-MRI capabilities existing in Slicer 2.x
Core infrastructure for DT-MRI processing and visualization, fiber processing and visualization
DTI Data Model
MRML nodes for different data representations involved in DTI analysis:
- Diffusion Weighted Images: vtkMRMLDiffusionWeightedVolumeNode.
- Diffusion Tensor Images: vtkMRMLDiffusionTensorVolumeNode.
- Fiber Bundles: vtkMRMLFiberBundleNode.
Visualization/Display
MRML nodes for definition for DTI data visualization:
- Diffusion Weighted Images: vtkMRMLDiffusionWeightedVolumeDisplayNode.
- Diffusion Tensor Images: vtkMRMLDiffusionTensorVolumeDisplayNode, and vtkMRMLDiffusionTensorDisplayPropertiesNode.
- Fiber Bundles: vtkMRMLFiberBundleDisplayNode, vtkMRMLFiberBundleLineDisplayNode, vtkMRMLFiberBundleTubeDisplayNode, and vtkMRMLFiberBundleGlyphDisplayNode.
Storage and I/O
- DWI and DTI I/O: NRRD is the format supported by Slicer 3 for storing DWI and DTI images.
- NNRD reader/writer: vtkNRRDReader and vtkNRRDWriter.
- Storage node: vtkMRMLNRRDStorageNode.
- Fiber I/O: vtkPolyData has been the format adopted for the description of fibers.
- Storage node: vtkMRMLFiberBundleStorageNode
Displaying Logic
- Visualization pipelines for DTI, DWI volumes, and fiber bundles are incorporated into the corresponding display nodes.
- DWI volumes are displayed as separate components.
- DTI volumes are displayed computed scalar properties (such as FA, Linear Measure, etc.)
- Fiber bundles are displayed as lines, tubes, and glyphs with their own properties and colors.
Diffusion Processing Toolbox
vtkTeem-library provides tools for:
- Tensor estimation
- Computation of scalar measurements from tensor fields
- Fast rendering of tensor fields using glyphs: line, box, ellipsoid.
- Fiber Tracking using integration techniques
- Multiple ROI seeding and logic interconnections between ROIs
DT-MRI processing and visualization Modules
Tensor Estimation from DWI
- CLI Module: DiffusionTensorEstimation.
Teem currently provides a clean interface to do this estimation in a voxel by voxel fashion. Collaboration with Gordon Kindlmann for a vtk filter implementation that encapsulates the estimation process (vtkTeemEstimateDiffusionTensor).
Diffusion Tensor Scalar Measurements
- CLI Module: DiffusionTensorMathematics.
- Implemented in vtkTeem library vtkDiffusionTensorMathematics.
Rician LMMSE Filter
- CLI Module: dwiNoiseFilter.
- Filters a set of diffusion weighted images in the mean squared error sense using a Rician noise model. The noise parameter is automatically estimated.
- Contributed by Santiago Aja Fernandez and Marc Niethammer
Tractography Display/Load/Save Module
- Load and Saves fiber tracks as vtkPolyData files (.vtp)
- Displays fiber tracks as lines, tubes, glyphs. Allows to control line/tube/glyph display properties and colors
- Built-in Slicer3 module.
Tractography ROI Seeding Module
- Creates fibers passing through a specified ROI
- CLI Module: ROI Seeding.
Tractography Fiducial Seeding Module
- Creates fibers passing through the specified fiducial points interactively
- Built-in Slicer3 module: vtkSlicerTractographyFiducialSeedingGUI.cxx.
Tractography ROI Select Module
- Select tracts passing or not passing through ROIs
- CLI Module: ROI Select.
Stochastic Tractography Filter
- CLI Module: StochasticTractographyFilter.
- Generates a map of connectivity probabilities from a DWI volume.).
- Contributed by Tri Ngo (tringo@gmail.com)
ROI Tract Filter
- CLI Module: ROITractFilter.
- Creates a new tract container containing only tracts which pass through the selected ROI's.
- Contributed by Tri Ngo (tringo@gmail.com)
Generate Connectivity Map
- CLI Module: GenerateConnectivityMap.
- Generates a volume where the value of each voxel is the number of fibers which pass through that voxel divided by the total number of sampled fibers. This value can been interpreted as the probability that a particular voxel is connected to the seed ROI by a fiber tract.
- Contributed by Tri Ngo (tringo@gmail.com)
Future development plans
- Fiber editing: enviroment for manually editing individual fibers/bundles, reassigning of fibers to bundles.
- Teem based tractography (Core 1 & 2).
- Fiber Bundle Clustering (Core 1).
- Render glyphs in the 2D slice windows.
- Statistics along fiber tracts (Core 1).
- Quantitative measurement
- Tract-based
- Region of interest-based
- fMRI seeding
- Surgical planning
- DT-MRI segmentation/atlas creation: enviroment for segmentation of DT-MRI fields
- DT-MRI registration: enviroment for registration of DT-MRI fields (possibly via DWI registration -- work done at GE and presented in MICCAI '06).
- Tensor estimation using different methods, namely:
- Least Squares
- Weighted Least Squares
- Non-linear methods
- Maximum Likelihood approach
Development Screenshots
Notes on general diffusion framework (ODF/2 tensor) support
http://wiki.na-mic.org/Wiki/index.php/Slicer3:DTMRI:GeneralDiffusionFramework