Slicer3:Diffusion Editor
From NAMIC Wiki
Home < Slicer3:Diffusion Editor
Contents
Overview
The goal of this project is to add a Gradient Editor for DWI data in the Volumes modul of Slicer3. As the documentation of gradients in dicom data is not standardized, MRI scanners handle them differently. Because of that there is a big need to add/modify gradients manually.
Interface
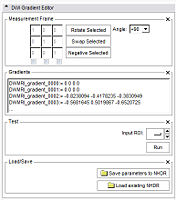
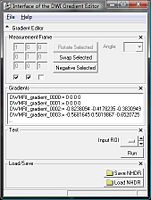
This screen shots show the interface and its changes during the last months.
- Interface
How to use the editor
- 1.Step: Generate a Nrrd-header of your DWI data. Use the "Dicom to Nrrd" module in Slicer3 (Modules->Converters->Dicom to Nrrd).
- -> A nhdr-file will be created.
- 2.Step: Use the Output (nhdr-file) as an Input of the Volumes module.
- -> The Gradient Editor will be enabled. If you load any other type of dataset the Editor will be disabled.
What it can (in a while)
- Choose Measurement Frame.
- Negative: Select the columns you want to negative.
- Swap: Select two columns you want to swap.
- Rotate: Select one column you want to rotate by an angle you can
- choose from a given set of values.
- set yourself.
- Enable Matrix: Set your own values.
- Define Gradients.
- If the .nhdr file has the information about the gradients, the editor will put them in the text field.
- If there is no information about the gradients, the GE gradients will be displayed by default.
- You can copy/paste your own gradients in the text field.
- You can load gradients from a plain text file or .nhdr file.
- Run test.
- Add a ROI label map. To see if the chosen parameters are reasonable, you can run a test that computes the Tractography Seeding.
- Save the parameters to the .nhdr file.
Additional Information
Nrrd format: http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:DTI:Nrrd_format