Difference between revisions of "Slicer3:Fluorescence and Electron Microscopy Support"
From NAMIC Wiki
| Line 1: | Line 1: | ||
Back to [[NA-MIC_Collaborations|NA-MIC_Collaborations]] | Back to [[NA-MIC_Collaborations|NA-MIC_Collaborations]] | ||
| − | + | = Name of Template project goes here = | |
| − | + | Analyze dendritic spine morphologies from electron tomographic reconstructions. | |
| − | + | = Description = | |
| − | |||
| − | |||
| + | Dendritic spines are the primary sites of excitatory neuronal communication in the brain. It is of fundamental interest to cellular neurobiologists to understand how the number, size, and shape of dendritic spines varies across brain regions, and as a function of experimental treatment within a given region. | ||
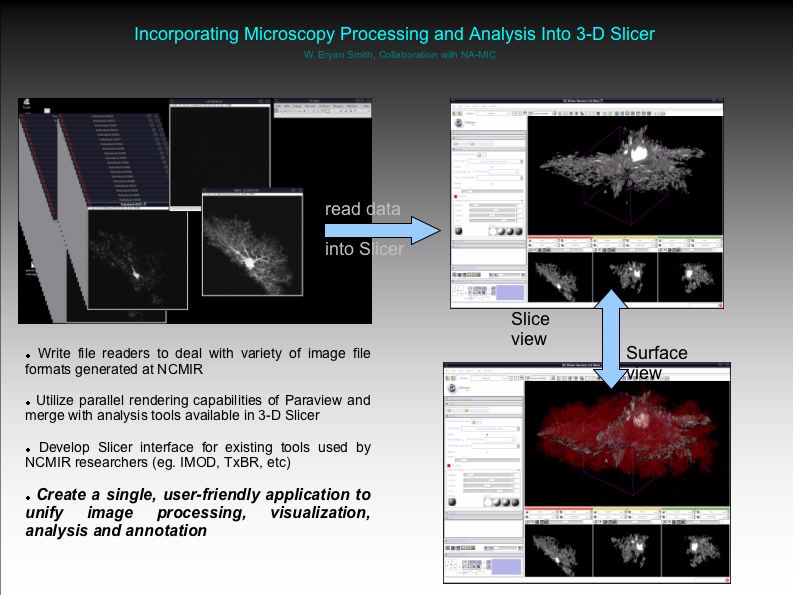
| − | + | ''Objective (Light Microscopy)'' Incorporate into Slicer our existing microscopy processing and analysis routines, currently being done in Matlab. | |
| − | + | ''Progress'' | |
Reading 3-D TIFF images and generating surface models has been easily accomplished. | Reading 3-D TIFF images and generating surface models has been easily accomplished. | ||
| Line 23: | Line 22: | ||
** Can ParaView be used as visualization tool for surface rendering? Is it faster? | ** Can ParaView be used as visualization tool for surface rendering? Is it faster? | ||
| − | + | = Key Investigators = | |
| + | |||
* NCMIR/UCSD: W. Bryan Smith, Mark Ellisman | * NCMIR/UCSD: W. Bryan Smith, Mark Ellisman | ||
* Isomics: Steve Pieper | * Isomics: Steve Pieper | ||
| − | + | = Links = | |
* A [[Media:NCMIR example dataset.zip | sample dataset]] is available. | * A [[Media:NCMIR example dataset.zip | sample dataset]] is available. | ||
| + | *[http://ncmir.ucsd.edu/~bryan/spines01.jpg A segmented spiny dendrite from cerebellum] | ||
| + | *[http://ncmir.ucsd.edu/~bryan/spineSample.jpg An example set of segmented spines] | ||
| − | |||
[[Image:SlicerAstrocyte.jpg | NCMIR image processing pipeline]] | [[Image:SlicerAstrocyte.jpg | NCMIR image processing pipeline]] | ||
Revision as of 19:05, 28 November 2007
Home < Slicer3:Fluorescence and Electron Microscopy SupportBack to NA-MIC_Collaborations
Name of Template project goes here
Analyze dendritic spine morphologies from electron tomographic reconstructions.
Description
Dendritic spines are the primary sites of excitatory neuronal communication in the brain. It is of fundamental interest to cellular neurobiologists to understand how the number, size, and shape of dendritic spines varies across brain regions, and as a function of experimental treatment within a given region.
Objective (Light Microscopy) Incorporate into Slicer our existing microscopy processing and analysis routines, currently being done in Matlab.
Progress Reading 3-D TIFF images and generating surface models has been easily accomplished.
- 2007-04-26: Modifying Otsu Segmentation CLM to make use of Connected Components Image Filter.
- Works in Linux, not OS X.
- 2007-04-25: Discussion of relevant steps to accomplish initial astrocyte parsing project.
- Using EM Segmentation or 2-D histogram normalization approach to segment immunostaining data.
- Need 3-D erosion functionality for 'shrinking' astrocyte image volume to separate into two domains: an outer, "hull", and an inner, "core" domain in which differential protein distributions will be assessed.
- Can ParaView be used as visualization tool for surface rendering? Is it faster?
Key Investigators
- NCMIR/UCSD: W. Bryan Smith, Mark Ellisman
- Isomics: Steve Pieper