Difference between revisions of "Slicer3:Fluorescence and Electron Microscopy Support"
From NAMIC Wiki
| Line 19: | Line 19: | ||
'''Links:''' | '''Links:''' | ||
| − | * Some sample data are available here: [[Image:NCMIR example dataset.zip | + | * Some sample data are available here: [[Image:NCMIR example dataset.zip]] |
'''Screenshot:''' | '''Screenshot:''' | ||
[[Image:SlicerAstrocyte.jpg]] | [[Image:SlicerAstrocyte.jpg]] | ||
Revision as of 21:04, 3 May 2007
Home < Slicer3:Fluorescence and Electron Microscopy SupportBack to NA-MIC_Collaborations
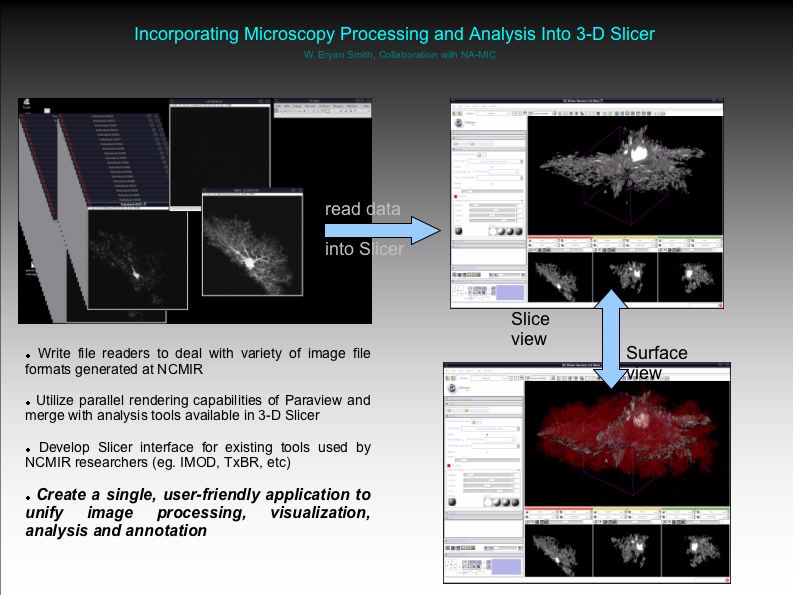
Objective: Incorporate into Slicer our existing microscopy processing and analysis routines, currently being done in Matlab.
Progress: Reading 3-D TIFF images and generating surface models has been easily accomplished.
- 2007-04-26: Modifying Otsu Segmentation CLM to make use of Connected Components Image Filter.
- Works in Linux, not OS X.
- 2007-04-25: Discussion of relevant steps to accomplish initial astrocyte parsing project.
- Using EM Segmentation or 2-D histogram normalization approach to segment immunostaining data.
- Need 3-D erosion functionality for 'shrinking' astrocyte image volume to separate into two domains: an outer, "hull", and an inner, "core" domain in which differential protein distributions will be assessed.
- Can ParaView be used as visualization tool for surface rendering? Is it faster?
Key Investigators:
- NCMIR/UCSD: W. Bryan Smith, Mark Ellisman
- Isomics: Steve Pieper
Links:
- Some sample data are available here: File:NCMIR example dataset.zip
Screenshot: