Slicer3:Slicer Daemon
Contents
Goals and Functionality
The Slicer Daemon refers to a network protocol that can be used to connect to a running instance of slicer to read and write data in the MRML scene and execute other commands. The name is based on the unix system convention of naming network services 'daemons'.
Server Implementation
The file slicerd.tcl implements the server side functionality.
By default it listens for connections on port 18943.
Clients
Tcl
Two utilities are provided:
- slicerget.tcl is used to read volumes out of slicer. The volume is written to the stdout of the slicerget command in nrrd format.
- slicerput.tcl is used to write volumes into slicer. The volume is read in nrrd format from stdin of slicerput and loaded into the mrml scene.
Some sample commands (assumes your PATH is correctly set to include unu, slicerget and slicerput):
# a noop -- just copy image onto itself slicerget.tcl 1 | slicerput.tcl noop
# put external data into slicer unu 1op abs -i d:/data/bunny-small.nrrd | slicerput.tcl
# run an external command and put the data back into slicer slicerget.tcl 1 | unu 1op abs -i - slicerput.tcl abs
Python
A Python based set of code for interacting with the Slicer Daemon is provided.
For example, the following code reads a volume and creates a new volume where each voxel is the square of the corresponding voxel of the input image. The new image is then sent back to slicer.
import slicerd import numpy s = slicerd.slicerd() n = s.get(0) im = n.getImage() n.setImage( im * im ) s.put(n, 'newImage')
For example, the following code reads a volume and extracts a slice of it for plotting using the matplotlib code (see the SciPy website for more info on Python numerics and plotting).
import slicerd import pylab s = slicerd.slicerd() n = s.get(0) slice = n.getImage()[16,:,:] pylab.imshow(slice) pylab.show()
Matlab
Note: this is initial documentation only, and is subject to change as the API evolves.
Matlab based versions of Slicer Daemon client code are available.
This project has been worked on during the NAMIC project week 2007.
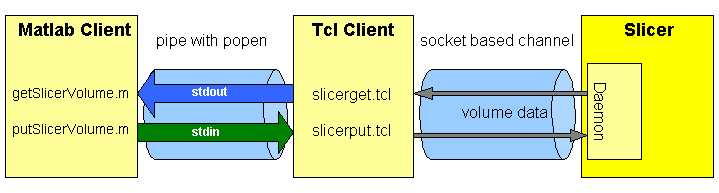
The Matlab scripts getSlicerVolume.m and putSlicerVolume.m use Matlab extention popen to connect to stdout(stdin respectively) of the tcl client slicerget.tcl (slicerput.tcl respectively). The tcl client establishes a channel to the SlicerDaemon socket and requests(sends) data.
The matlab toolbox in combination with the SlicerDaemon support the exchange of scalar and tensor volumes that are in orientation right-anterior-superior or left-posterior-superior. DWI volumes and other orientations are not supported yet.
Basic Slicer-Matlab tutorial
Here a short step by step tutorial how to send a volume first from Slicer to Matlab and then from Matlab abck to Slicer:
- Start Slicer3 with parameter "--daemon" and load the (scalar or tensor) volume you want to send to Matlab.
- Start Matlab
- In Matlab, just for conveniance, change into the "Matlab" subdirectory of the SlicerDaemon module in Slicer3 (something like ../Slicer3/Modules/SlicerDaemon/Matlab)
- Initally, the popen C functions need to be compiled in Matlab for your machine (this is not handled by cmake yet). This needs to be done only once:
mex popen/popenw.c mex popen/popenr.c
- Typing the following commang in Matlab, a Slicer volume with name "wcase1.nhdr" will be piped into a Matlab structure called "slicer_volume":
slicer_volume = getSlicerVolume('wcase1.nhdr')
The matlab strucuture for instance looks like this (for a scalar volume):
slicer_volume =
content: 'wcase1.nhdr'
type: 'short'
dimension: 3
space: 'right-anterior-superior'
sizes: [256 256 124]
endian: 'little'
encoding: 'raw'
spaceorigin: [119.5310 -92.2500 119.5310]
spaceunits: {'mm' 'mm' 'mm'}
kinds: {'space' 'space' 'space'}
data: [256x256x124 int16]
spacedirections: [3x3 double]
or like this (for a tensor volume):
slicer_volume =
content: 'helix.nhdr'
type: 'float'
dimension: 4
space: 'right-anterior-superior'
sizes: [7 64 32 12]
endian: 'little'
encoding: 'raw'
spaceorigin: [-6.9386 -28.7554 -8.7247]
spaceunits: {'"mm"' '"mm"' '"mm"'}
kinds: {'3D-masked-symmetric-matrix' 'space' 'space' 'space'}
data: [4-D single]
spacedirections: [3x3 double]
measurementframe: [3x3 double]
centerings: {'???' 'cell' 'cell' 'cell'}
All volumes that come out of Slicer are in 'right-anterior-superior' orientation, have 'raw' encoding, and 'little' endian. Even if the original file loaded into Slicer had other header parameters.
- Now the volume data can be processed in Matlab. Just for example, the volume can be thresholded:
slicer_volume.data(slicer_volume.data > 100) = 0;
By changing the field "content", the name of the volume node in Slicer will be changed:
slicer_volume.content='Matlab_says_hi';
- This is the command to send the volume back to Slicer:
putSlicerVolume(slicer_volume)