Slicer:Feedback
From NAMIC Wiki
Home < Slicer:Feedback
3D Slicer is a free open source software package distributed under a BSD style license. The majority of funding for the development of 3D slicer comes from a number of grants and contracts from the National Institutes of Health (see slicer acknowledgements for more information).
We are currently preparing an application to extend funding for one of the major grants that contribute to this effort.
We are looking to include information about the impact that slicer has had. Specifically, we are looking for peer reviewed journal papers and funded grants which rely on 3D Slicer. Please email us or add to the bottom of this page.
Peer reviewed journal papers:
- AJNR Am J Neuroradiol. 2006 May;27(5):1032-6.
- A method for clustering white matter fiber tracts.
- O'Donnell LJ, Kubicki M, Shenton ME, Dreusicke MH, Grimson WE, Westin CF.
- MIT Computer Science and AI Lab, Cambridge, MA 02139, USA.
- BACKGROUND/PURPOSE: Despite its potential for visualizing white matter fiber tracts in vivo, diffusion tensor tractography has found only limited applications in clinical research in which specific anatomic connections between distant regions need to be evaluated. We introduce a robust method for fiber clustering that guides the separation of anatomically distinct fiber tracts and enables further estimation of anatomic connectivity between distant brain regions. METHODS: Line scanning diffusion tensor images (LSDTI) were acquired on a 1.5T magnet. Regions of interest for several anatomically distinct fiber tracts were manually drawn; then, white matter tractography was performed by using the Runge-Kutta method to interpolate paths (fiber traces) following the major directions of diffusion, in which traces were seeded only within the defined regions of interest. Next, a fully automatic procedure was applied to fiber traces, grouping them according to a pairwise similarity function that takes into account the shapes of the fibers and their spatial locations. RESULTS: We demonstrated the ability of the clustering algorithm to separate several fiber tracts which are otherwise difficult to define (left and right fornix, uncinate fasciculus and inferior occipitofrontal fasciculus, and corpus callosum fibers). CONCLUSION: This method successfully delineates fiber tracts that can be further analyzed for clinical research purposes. Hypotheses regarding specific fiber connections and their abnormalities in various neuropsychiatric disorders can now be tested.
- Grant Support:
- 1-R01-NS051826-01/NS/NINDS
- K02 MH 01110/MH/NIMH
- P41 RR13218/RR/NCRR
- R03 MH 068464-02/MH/NIMH
- U54 EB005149/EB/NIBIB
- your paper here
Peer reviewed exhibits:
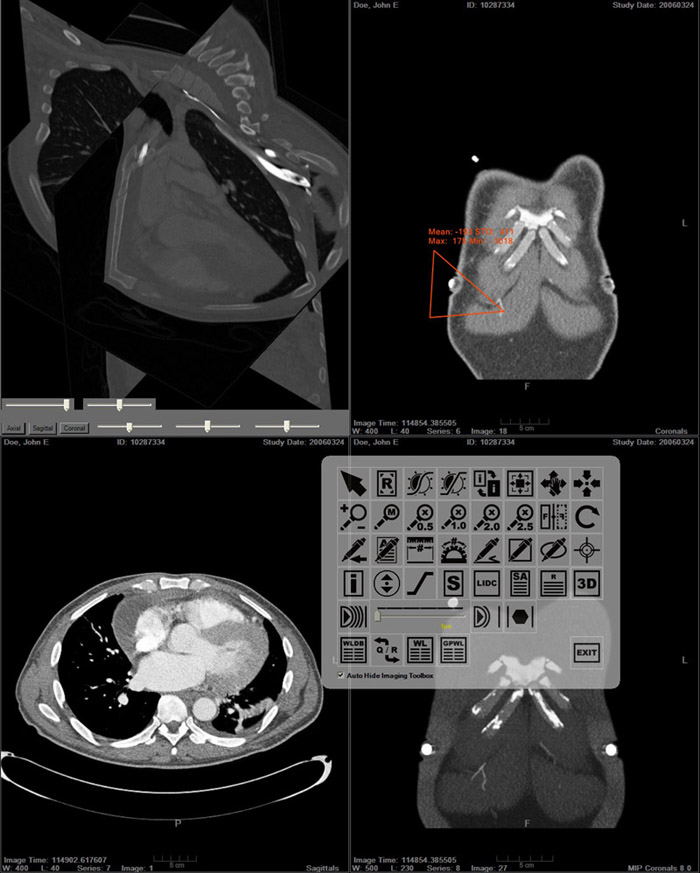
- RSNA 2006.
- A Translation Station for Imaging
- P Mongkolwat, PhD; T Lechner, MS; A Kogan; S Talbot; B Johnson, OD; D S Channin, MD
- Imaging Informatics Section, Northwestern University, Chicago, IL 60611, USA
- Abstract: while providing clinical feedback to the basic sciences. In imaging informatics, this means enabling collaborations between radiologists and computer scientists. Commercial workstations do not allow for experimentation nor do they evolve fast enough. Research imaging packages, while innovative, do not provide for clinical activities or regulatory compliance. We added IHE workflow functionality, hanging protocols and other clinical functions to the SLICER research platform. We leveraged the functionality and methodology of the Insight and Visualization Toolkits, and the DCM4CHE tools. A research mode allows for the evaluation of experimental algorithms against a PACS while protecting patient confidentiality. This combined functionality allows researchers to develop algorithms against clinical cases while at the same allowing clinicians to explore and validate these developments.