Summer2009:Registration reproducibility in Slicer
Key Investigators
- BWH: Andriy Fedorov, Steve Pieper, Tina Kapur
- GE: Jim Miller
- Kitware: Luis Ibanez
- EAB: Bill Lorensen
Objective
Rigid registration in general, and RigidRegistration module of Slicer3 in particular, are important workflow components for a number of applications. Two issues are of our interest:
- We found gross inconsistency between the result of using rigid registration module on volumetric brain MRI in GUI vs command line invocation.
- There is also significant difference between command line invocations of the module on different platforms.
The issue has been confirmed independently by Andriy, Bill, Jim and Steve. It was also confirmed by Kilian. The problem is thoroughly documented in Slicer3 bug 416.
We would like to understand the source of this inconsistency, together with the understanding what is reasonable to expect expect in terms of reproducibility from such complex numerical codes like rigid registration.
Approach, Plan
- use clinical data to demonstrate the problem, once again
- collect feedback from registration experts
- create a test that demonstrates the problem, and add it to the ITK dashboard
- Questions to consider:
- is reproducibility a function of the architecture? is it dependent on multi-threading? can it be quantified and estimated?
- what is the correct way to test reproducibility for this kind of applications?
Progress
- added testing mode to RigidRegistration to measure the maximum difference
- abused Slicer3 dashboard to collect the magnitude of difference on various platforms
Test setup
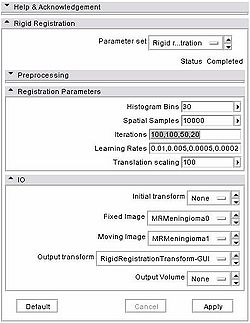
1. Get images and transform from Slicer3 trunk:
2. To get the transform in GUI, go to RigidRegistration module, use the default parameters to initialize (see figure), EXCEPT reduce iteration cout from the default to "100,100,50,20" 3. To get the transform with CLI, here's the command line you need to run: Slicer3 --launch RigidRegistration --fixedsmoothingfactor 0 --movingsmoothingfactor 0 --histogrambins 30 --spatialsamples 10000 --iterations 100,100,50,20 --learningrate 0.01,0.005,0.0005,0.0002 --translationscale 100 --outputtransform ./RigidRegistrationTransform-CLI.txt MRMeningioma0.nrrd MRMeningioma1.nrrd 4. To get the actual difference in the corner of your image using one transform vs another, you can run RigidRegistration in the testing mode: |