Difference between revisions of "User:Haehn"
| (12 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
! Info || | ! Info || | ||
|- style="vertical-align:top" | |- style="vertical-align:top" | ||
| − | |'''Daniel | + | |'''Daniel Haehn''' |
| + | |||
| + | http://danielhaehn.com | ||
| + | |||
| + | || <center>[[Image:haehn.jpg]]</center> | ||
|} | |} | ||
| − | |||
| − | + | === VMTK in 3D Slicer === | |
| − | + | ||
| + | The Vascular Modeling Toolkit ([http://vmtk.org VMTK]) is a collection of libraries and tools for 3D reconstruction, geometric analysis, mesh generation and surface data analysis for image-based modeling of blood vessels. It should be very interesting to offer such techniques in 3D Slicer. | ||
| + | |||
| + | The official project page: | ||
| + | http://www.vmtk.org/Main/VmtkIn3DSlicer | ||
| − | |||
| − | + | More recent news on this project: <br> | |
| − | + | http://www.na-mic.org/Wiki/index.php/2009_Winter_Project_Week_Slicer_VMTK<br> | |
| + | http://www.na-mic.org/Wiki/index.php/Summer2009:The_Vascular_Modeling_Toolkit_in_3D_Slicer<br> | ||
| + | http://www.na-mic.org/Wiki/index.php/2010_Winter_Project_Week_The_Vascular_Modeling_Toolkit_in_3D_Slicer | ||
| + | http://www.na-mic.org/Wiki/index.php/2010_Summer_Project_Week/The_Vascular_Modeling_Toolkit_in_3D_Slicer | ||
| + | http://www.na-mic.org/Wiki/index.php/2011_Winter_Project_Week:The_Vascular_Modeling_Toolkit_in_3D_Slicer | ||
| + | http://www.na-mic.org/Wiki/index.php/2011_Winter_Project_Week:StenosisDetector | ||
| − | + | This work is done in conjunction with Luca Antiga (Medical Imaging Unit, Bioengineering Department, Mario Negri Institute, Bergamo, Italy). | |
| − | === | + | ==== VMKT in Slicer4 ==== |
| − | + | The VMTK integration in 3D Slicer will be transitioned to Slicer4. | |
| − | + | Some developer notes: | |
| + | <pre> | ||
| + | # clone the fork | ||
| + | git clone git@github.com:haehn/vmtk.git | ||
| − | + | # check the status | |
| + | git status | ||
| − | + | # switch to branch superbuild | |
| + | git checkout superbuild | ||
| + | |||
| + | # include upstream by editing .git/config | ||
| + | # 17 [remote "upstream"] | ||
| + | # 18 url = https://github.com/lantiga/vmtk.git | ||
| + | # 19 fetch = +refs/heads/*:refs/remotes/upstream/* | ||
| + | |||
| + | # get the upstream | ||
| + | git fetch upstream | ||
| + | |||
| + | # merge the latest changes to local | ||
| + | git merge upstream/superbuild | ||
| + | |||
| + | # push the latest changes from upstream to fork | ||
| + | git push | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | Now, somebody forked the repo, made some changes we want in the master: | ||
| + | |||
| + | <pre> | ||
| + | # add the remote repo | ||
| + | git remote add jcfr https://github.com/jcfr/EMSegment4.git | ||
| + | |||
| + | # fetch it | ||
| + | git fetch jcfr | ||
| + | |||
| + | # see diff | ||
| + | git log -p master jcfr/master | ||
| + | |||
| + | # now merge the remote with our master | ||
| + | git merge jcfr/master | ||
| + | Updating f5d1b39..dd72623 | ||
| + | error: Your local changes to 'CMakeLists.txt' would be overwritten by merge. Aborting. | ||
| + | Please, commit your changes or stash them before you can merge. | ||
| + | |||
| + | # let's revert our local changes | ||
| + | git checkout CMakeLists.txt | ||
| + | |||
| + | # merge again | ||
| + | git merge jcfr/master | ||
| + | |||
| + | # and push it to the master | ||
| + | git push | ||
| + | </pre> | ||
| + | |||
| + | <pre> | ||
| + | # use cherry-pick to merge a commit from another branch into the current branch | ||
| + | git cherry-pick COMMITHASH | ||
| + | |||
| + | # now push | ||
| + | git push | ||
| + | </pre> | ||
| + | |||
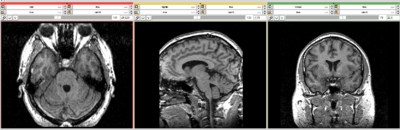
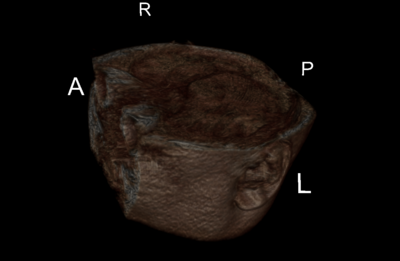
| + | === Images === | ||
| + | {| | ||
| + | |[[File:Becky.png|thumb|400px]]||[[File:Becky2.png|thumb|400px]]||[[File:Beck3.png|thumb|400px]] | ||
| + | |} | ||
Latest revision as of 14:31, 14 March 2013
Daniel Haehn's Page
| Info | |
|---|---|
| Daniel Haehn |  |
VMTK in 3D Slicer
The Vascular Modeling Toolkit (VMTK) is a collection of libraries and tools for 3D reconstruction, geometric analysis, mesh generation and surface data analysis for image-based modeling of blood vessels. It should be very interesting to offer such techniques in 3D Slicer.
The official project page: http://www.vmtk.org/Main/VmtkIn3DSlicer
More recent news on this project:
http://www.na-mic.org/Wiki/index.php/2009_Winter_Project_Week_Slicer_VMTK
http://www.na-mic.org/Wiki/index.php/Summer2009:The_Vascular_Modeling_Toolkit_in_3D_Slicer
http://www.na-mic.org/Wiki/index.php/2010_Winter_Project_Week_The_Vascular_Modeling_Toolkit_in_3D_Slicer
http://www.na-mic.org/Wiki/index.php/2010_Summer_Project_Week/The_Vascular_Modeling_Toolkit_in_3D_Slicer
http://www.na-mic.org/Wiki/index.php/2011_Winter_Project_Week:The_Vascular_Modeling_Toolkit_in_3D_Slicer
http://www.na-mic.org/Wiki/index.php/2011_Winter_Project_Week:StenosisDetector
This work is done in conjunction with Luca Antiga (Medical Imaging Unit, Bioengineering Department, Mario Negri Institute, Bergamo, Italy).
VMKT in Slicer4
The VMTK integration in 3D Slicer will be transitioned to Slicer4.
Some developer notes:
# clone the fork git clone git@github.com:haehn/vmtk.git # check the status git status # switch to branch superbuild git checkout superbuild # include upstream by editing .git/config # 17 [remote "upstream"] # 18 url = https://github.com/lantiga/vmtk.git # 19 fetch = +refs/heads/*:refs/remotes/upstream/* # get the upstream git fetch upstream # merge the latest changes to local git merge upstream/superbuild # push the latest changes from upstream to fork git push
Now, somebody forked the repo, made some changes we want in the master:
# add the remote repo git remote add jcfr https://github.com/jcfr/EMSegment4.git # fetch it git fetch jcfr # see diff git log -p master jcfr/master # now merge the remote with our master git merge jcfr/master Updating f5d1b39..dd72623 error: Your local changes to 'CMakeLists.txt' would be overwritten by merge. Aborting. Please, commit your changes or stash them before you can merge. # let's revert our local changes git checkout CMakeLists.txt # merge again git merge jcfr/master # and push it to the master git push
# use cherry-pick to merge a commit from another branch into the current branch git cherry-pick COMMITHASH # now push git push