Validation and testing of 3D Slicer modules implementing the Utah segmentation algorithm for traumatic brain injury
From NAMIC Wiki
Home < Validation and testing of 3D Slicer modules implementing the Utah segmentation algorithm for traumatic brain injury
Key Investigators
- Utah: Bo Wang, Marcel Prastawa, Guido Gerig
- UCLA: Andrei Irimia, Micah Chambers, Jack Van Horn
Objective
- The primary objective is for the Utah team to coordinate with UCLA regarding the release of a Slicer module to perform multimodal automatic segmentation of TBI using the methodologies developed by the Utah team.
- One planned topic of interest includes a progress update to the UCLA group on the current status of the matlab/C++/VTK translation of the Utah algorithm
- We intend to spend some time on testing the method as well as on getting feedback on the best way to integrate the algorithm within Slicer
- It would also be useful to demonstrate how the algorithm performs on the UCLA data which have already been made available to the Utah team
Approach, Plan
- Bo and Marcel will be working together on the C++ implementation of the algorithm.
- Micah and Bo discuss the workflow and user interface in Slicer.
- Set up svn (GitHub & NITRC) for the development of this project.
- Test and debug the preliminary C++ code.
- Try to build the command-line module.
- Discuss with our collaborators from UCLA about their requirements.
Progress
- We've set up new accounts at GitHub and NITRC for this project which is called 4D-PARSeR (Pathological Anatomy Regression via Segmentation and Registration), and the current C++ code has been committed to svn.
- The status of current C++ implementation is that all the major components are done. The major components include intra-time-point alignment, atlas alignment to each time point, appearance model (density estimation), and basic deformable registration.
- Micah and Bo had discussed the workflow and the user interface of the algorithm. Micah discussed with Slicer experts about the possible solutions for the user interface of longitudinal multimodal images. About the UI, Ron recommended us to use the Longitudinal PET/CT Analysis module in Slicer as a reference, and then Micah talked to Andrey about the UI of the Longitudinal PET/CT Analysis module.
- We've been working on testing and debugging the current C++ code together. Bo tested loading and saving segmentation results, saving registration deformation field.
- Learned to use the new Slicer deformation field visualization module for visualizing the deformation field in different ways(such as arrows, grid, and isosurface).
- Talked to several experts (Peter, Lauren, and Demian) working on both diffusion tensor and structural MR images to learn how to process and analyze both diffusion tensor and structural MR images together.
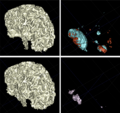
- The segmentation results of 4D-PARSeR has been used in joint DTI and MRI analysis of TBI patients.
Delivery Mechanism
- Slicer module
- Standalone module (command line) for the first step
References
- TBI DBP
- Bo Wang, Marcel Prastawa, Andrei Irimia, Micah C. Chambers, Neda Sadeghi, Paul M. Vespa, John D. Van Horn, Guido Gerig, Analyzing Imaging Biomarkers for Traumatic Brain Injury Using 4D Modeling of Longitudinal MRI, In IEEE ISBI 2013 .
- Bo Wang, Marcel Prastawa, Suyash P. Awate, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn, Guido Gerig, Segmentation of Serial MRI of TBI patients using Personalized Atlas Construction and Topological Change Estimation, In Proceedings of IEEE ISBI 2012, pp. 1152--1155. 2012.