Difference between revisions of "Events:August 2008 NCBC AHM"
(→Tools) |
(→Agenda) |
||
| (28 intermediate revisions by 6 users not shown) | |||
| Line 7: | Line 7: | ||
* Wednesday - August 13th, 8:30 AM - 6:30 PM | * Wednesday - August 13th, 8:30 AM - 6:30 PM | ||
** 8:30 - 8:45 AM Introduction by Dr. Zerhouni, or Dr Berg, Natcher Auditorium | ** 8:30 - 8:45 AM Introduction by Dr. Zerhouni, or Dr Berg, Natcher Auditorium | ||
| − | ** 8:45 - 10:15 AM NCBC Presentations from PIs ('''Ron''', 10 | + | ** 8:45 - 10:15 AM NCBC Presentations from PIs ('''Ron''', [[media:NA-MIC 2008.ppt|10 minute overview over NA-MIC]]) |
** 10:15 - 10:30 AM Big P and Building Bridges Summary (Russ Altman) | ** 10:15 - 10:30 AM Big P and Building Bridges Summary (Russ Altman) | ||
** 10:30 - 11:00 AM Break | ** 10:30 - 11:00 AM Break | ||
***PI Meeting with Drs. Zerhouni, Berg, Lindberg, Natcher A | ***PI Meeting with Drs. Zerhouni, Berg, Lindberg, Natcher A | ||
| − | ** 11:00 - 12:15 PM Presentations from Jr. Faculty Natcher Auditorium ('''Tom Fletcher''': 10 Minutes) | + | ** 11:00 - 12:15 PM Presentations from Jr. Faculty Natcher Auditorium ('''Tom Fletcher''': 10 Minutes - [[media:Fletcher_NAMIC_NCBC2008.ppt|"Statistical Analysis of Anatomy from Medical Images"]]) |
** 12:15 - 1:30 PM Working Lunch: Working Groups and Building Bridges | ** 12:15 - 1:30 PM Working Lunch: Working Groups and Building Bridges | ||
| − | ** 1:30 - 3:30 PM "Hot Topic" Presentations Natcher Auditorium (30 Mins - '''Ross Whitaker''' - "NAMIC Highlights: From Algorithms and Software to Biomedical Science"): The best science of the center, focus on Biology impact and DBPs. Share the highlights/titles by July 15th. | + | ** 1:30 - 3:30 PM "Hot Topic" Presentations Natcher Auditorium (30 Mins - '''Ross Whitaker''' - [[media:namic_highlights_aug08.ppt|"NAMIC Highlights: From Algorithms and Software to Biomedical Science"]], [[media:namic_highlights_aug08.pdf|pdf-version]]): The best science of the center, focus on Biology impact and DBPs. Share the highlights/titles by July 15th. |
** 3:30 - 3:45 PM Break | ** 3:30 - 3:45 PM Break | ||
** 3:45 - 5:15 PM "Hot Topic" Presentations continued | ** 3:45 - 5:15 PM "Hot Topic" Presentations continued | ||
| Line 19: | Line 19: | ||
** 6:30 - --- PM Open Reception at hotel with Industry Host. PIs and Project Team will join at 6:45PM. PI Meeting with Project team, Natcher A | ** 6:30 - --- PM Open Reception at hotel with Industry Host. PIs and Project Team will join at 6:45PM. PI Meeting with Project team, Natcher A | ||
* Thursday - August 14th, 8:30 AM - 12:15 PM | * Thursday - August 14th, 8:30 AM - 12:15 PM | ||
| − | ** 8:30 - 9:30 AM Presentation from each Center: Introduction to the science fair program demos and schedule (teaser for the Science Fair, '''Will Schroeder''') | + | ** 8:30 - 9:30 AM Presentation from each Center: Introduction to the science fair program demos and schedule ([[media:NAMICScienceFairTeaser.ppt |"teaser for the Science Fair"]], '''Will Schroeder''') |
| − | ** 9:30 - 11:00 AM | + | ** 9:30 - 11:00 AM Science Fair: [[media:NA-MIC kit NCBC AHM2008 SPujol.ppt |"Three ways to use the NA-MIC kit"]] ('''Sonia Pujol''') |
| + | Abstract: The National Alliance for Medical Imaging Computing (NA-MIC) develops a collection of tools and applications to improve the deployment of advanced image analysis methods to the research community. By combining science and technology, the NA-MIC kit fosters leading edge clinical research achievements in a collaborative and open-source environment. | ||
| + | The Science Fair will present the NA-MIC kit through the perspectives of each of our three target audiences: clinical researchers, biomedical engineers and algorithm developers. Attendees will be exposed to potential uses of the NA-MIC kit in the context of 3D visualization to facilitate data interpretation, in the use of advanced image analysis to extract relevant information, and in the development of tailored plug-ins to extend image computing capabilities. Through demonstrations of image processing methods within the 3D Slicer deployment platform, and presentations of NA-MIC pragmatic teaching initiatives to translate techniques into skills, the Science Fair will provide an overview of the benefits of NA-MIC efforts to the research community. | ||
** 11:00 - 12:00 noon Working Groups Report Out, Future Plans, Discussion(s) | ** 11:00 - 12:00 noon Working Groups Report Out, Future Plans, Discussion(s) | ||
** 12:00 - 12:15 PM Wrap-up and adjournment of main program (Conference ends for all but PIs, and Project Team and invited guests) | ** 12:00 - 12:15 PM Wrap-up and adjournment of main program (Conference ends for all but PIs, and Project Team and invited guests) | ||
** 12.15 - 1.15 PM Lunch (Conference ends for all but PIs) DBP discussion: Diabetes, MH, others | ** 12.15 - 1.15 PM Lunch (Conference ends for all but PIs) DBP discussion: Diabetes, MH, others | ||
** 1.15 - 3:00 PM Moderated Discussions with other NIH Programs: PIs: CaBIG, CTSA, BIRN, NIH Clinical Center, others | ** 1.15 - 3:00 PM Moderated Discussions with other NIH Programs: PIs: CaBIG, CTSA, BIRN, NIH Clinical Center, others | ||
| − | | style="background: #ebeced" colspan="2" align="center"| <b>NA-MIC is one of | + | | style="background: #ebeced" colspan="2" align="center"| <b>NA-MIC is one of seven National Centers for Biomedical Computing, part of the NIH roadmap initiative.</b> |
<br>See [http://meetings.nigms.nih.gov/index.cfm?event=home&ID=4095 here] for the program of this years' all hands meeting. | <br>See [http://meetings.nigms.nih.gov/index.cfm?event=home&ID=4095 here] for the program of this years' all hands meeting. | ||
|- | |- | ||
| Line 36: | Line 38: | ||
==DBPs== | ==DBPs== | ||
A short summary of the original and renewal DBPs (this will also be used in the DBP sig breakout session). Basically title and one paragraph description. | A short summary of the original and renewal DBPs (this will also be used in the DBP sig breakout session). Basically title and one paragraph description. | ||
| + | |||
1) During the first three years of NA-MIC, Core 3 consisted of four DBPs which were grouped into two thrusts. | 1) During the first three years of NA-MIC, Core 3 consisted of four DBPs which were grouped into two thrusts. | ||
*Thrust 1 was directed by Drs. Shenton and Saykin. Thrust 2 was directed by Drs. Potkin and Kennedy. The focus of the research was to utilize neuroimaging tools to evaluate fronto-temporal connectivity abnormalities in schizophrenia, as well as abnormalities in hemispheric connections (i.e., corpus callosum), and abnormalities in the anterior limb of the internal capsule. Improved segmentation techniques, coregistration of structural MRI, DTI-MR, and fMRI, as well as novel processing tools for evaluating white matter fiber tracts and interregional functional connectivity were needed to accomplish these goals, and they were developed in conjunction with Cores 1 and 2. | *Thrust 1 was directed by Drs. Shenton and Saykin. Thrust 2 was directed by Drs. Potkin and Kennedy. The focus of the research was to utilize neuroimaging tools to evaluate fronto-temporal connectivity abnormalities in schizophrenia, as well as abnormalities in hemispheric connections (i.e., corpus callosum), and abnormalities in the anterior limb of the internal capsule. Improved segmentation techniques, coregistration of structural MRI, DTI-MR, and fMRI, as well as novel processing tools for evaluating white matter fiber tracts and interregional functional connectivity were needed to accomplish these goals, and they were developed in conjunction with Cores 1 and 2. | ||
| Line 41: | Line 44: | ||
2) Starting with the 4th year of NA-MIC, the DBPs were shifted from schizophrenia to lupus, autism, velocardiofacial syndrome (VCSF), and prostate cancer. | 2) Starting with the 4th year of NA-MIC, the DBPs were shifted from schizophrenia to lupus, autism, velocardiofacial syndrome (VCSF), and prostate cancer. | ||
| − | *These DBPs now drive the computational research within NA-MIC. Specifically: Drs. Jeremy Bockholt and Charles Gasparovi at the MIND Institute and the University of New Mexico are analyzing brain lesions in Neuropsychiatric Systemic Lupus Erythematosis, Drs. Heather Hazlett and Joseph Piven at University of North Carolina, Chapel Hill are conducting a longitudinal MRI study of early brain development in Autism, Dr. Marek Kubicki at Harvard Medical School is investigating VCSF as a genetic model for schizophrenia, and Dr. Gabor Fichtinger at Queens University is developing a robotic percutaneous surgery system for treatment of prostate cancer. | + | *These DBPs now drive the computational research within NA-MIC. Specifically: |
| + | ** Drs. Jeremy Bockholt and Charles Gasparovi at the MIND Institute and the University of New Mexico are analyzing brain lesions in Neuropsychiatric Systemic Lupus Erythematosis, | ||
| + | ** Drs. Heather Hazlett and Joseph Piven at University of North Carolina, Chapel Hill are conducting a longitudinal MRI study of early brain development in Autism, | ||
| + | ** Dr. Marek Kubicki at Harvard Medical School is investigating VCSF as a genetic model for schizophrenia, and | ||
| + | ** Dr. Gabor Fichtinger at Queens University is developing a robotic percutaneous surgery system for treatment of prostate cancer. | ||
[[Image:NA-MIC-Image-Gallery-2008.png|thumb|250px|[http://www.na-mic.org/pages/Special:PubDB_Gallery?collection=12 The NA-MIC image gallery]]] | [[Image:NA-MIC-Image-Gallery-2008.png|thumb|250px|[http://www.na-mic.org/pages/Special:PubDB_Gallery?collection=12 The NA-MIC image gallery]]] | ||
| Line 47: | Line 54: | ||
==Publications== | ==Publications== | ||
1) A few key journal publications that have emerged directly from each center (say the top 5 per center). These should not be presented in a center-specific context but just to show the volume and quality of scientific production. You should be able to get these directly from your progress reports. | 1) A few key journal publications that have emerged directly from each center (say the top 5 per center). These should not be presented in a center-specific context but just to show the volume and quality of scientific production. You should be able to get these directly from your progress reports. | ||
| − | * We | + | * We use our publications database to gather download statistics about our publications. See [http://www.na-mic.org/pages/Special:PubDB_Stats?collectionid=12 here] for an up-to-date view. |
*This are the top five papers from our database in terms of number of downloads between September 2007 and July 2008 | *This are the top five papers from our database in terms of number of downloads between September 2007 and July 2008 | ||
| − | *# http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=609 | + | *# [http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=609 Pohl K, Fisher J, Bouix S, Shenton M, McCarley R, Grimson E, Kikinis R, Wells W. Using the logarithm of odds to define a vector space on probabilistic atlases. Med Image Anal. 2007 Oct;11(5):465-77.] |
| − | *# http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=996 | + | *# [http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=996 Hata N, Piper S, Jolesz F, Tempany C, Black P, Morikawa S, Iseki H, Hashizume M, Kikinis R. Application of Open Source Image Guided Therapy Software in MR-guided Therapies. Int Conf Med Image Comput Comput Assist Interv. 2007;10(Pt 1):491-8] |
| − | *# http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=544 | + | *# [http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=544 Yeo B, Sabuncu M, Desikan R, Fischl B, Golland P. Effects of Registration Regularization and Atlas Sharpness on Segmentation Accuracy. Int Conf Med Image Comput Comput Assist Interv. 2007;10(Pt 1):683-91] |
| − | *# http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=361 | + | *# [http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=361 Bouix S, Martin-Fernandez M, Ungar L, Nakamura M, Koo M, McCarley R, Shenton M. On evaluating brain tissue classifiers without a ground truth. Neuroimage. 2007 Jul 15;36(4):1207-1224] |
| − | *# http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=540 | + | *# [http://www.na-mic.org/pages/Special:PubDB_View?dspaceid=540 Georgiou T, Michailovich O, Rathi Y, Malcolm J, Tannenbaum A. Distribution Metrics and Image Segmentation. Linear Algebra and its Applications. 2007;425(2-3):663-672] |
2) Some statistics of publications: total number in the last year or cumulative. Again, these should be presented in aggregate rather than by center. | 2) Some statistics of publications: total number in the last year or cumulative. Again, these should be presented in aggregate rather than by center. | ||
| Line 70: | Line 77: | ||
*Newsletters, conferences, and other activities promoted by the centers. | *Newsletters, conferences, and other activities promoted by the centers. | ||
**The semi-annual hands-on event called the "NA-MIC Project Week". | **The semi-annual hands-on event called the "NA-MIC Project Week". | ||
| + | |||
| + | ==Collaborations== | ||
| + | |||
| + | http://wiki.na-mic.org/Wiki/index.php/NA-MIC_External_Collaborations | ||
| + | |||
| + | ==Buiding Bridges Postdoc== | ||
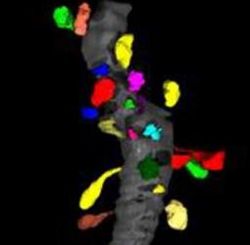
| + | [[Image:Spines01.jpg|thumb|250px|A small fragment of a dendrite (gray) with segmented spines (color).]] | ||
| + | * Who was the bridge built to? | ||
| + | **The National Center for Microscopy and Imaging Research (NCMIR; http://ncmir.ucsd.edu) at the University of California, San Diego. A key emphasis of NCMIR is the application of advanced imaging technologies to the nervous system in health and disease. Affiliated with UCSD’s Center for Research in Biological Systems (CRBS), the NCMIR is a recognized authority in the development of technologies for high throughput multi-scale imaging and analysis of biological systems at the mesoscale, the dimensional range spanning 5 nm3 and 50µm3. Macromolecules, organelles, and multi-component structures like synapses which are encompassed in this dimensional range have traditionally been challenging to study because they fall in the resolution gap between X-ray crystallography at one end and medical imaging at the other. | ||
| + | * What is the general topic of the research? | ||
| + | **Work within the Slicer environment to address multi-scale microscopy imaging data, thereby allowing for the analysis and visualization of cellular data. The project has focused on the importation of data into Slicer and the investigation of algorithms for cellular and sub-cellular data (e.g. see - http://www.na-mic.org/Wiki/index.php/Projects/Slicer3/2007_Project_Week_Support_for_electron_microscopy). | ||
| + | |||
| + | ==NA-MIC Description== | ||
| + | |||
| + | NA-MIC is a multi-institutional, interdisciplinary team of computer scientists, software engineers, and medical investigators who develop computational tools for the analysis and visualization of medical image data. The purpose of the center is to provide the infrastructure and environment for the development of computational algorithms and open source technologies, and then oversee the training and dissemination of these tools to the medical research community. This world-class software and development environment serves as a foundation for accelerating the development and deployment of computational tools that are readily accessible to the medical research community. The team combines cutting-edge computer vision research (to create medical imaging analysis algorithms) with state of the art software engineering techniques (based on "extreme" programming techniques in a distributed, open-source environment) to enable computational examination of both basic neuroscience and neurological disorders. In developing this infrastructure resource, the team is significantly expanding upon proven open systems technology and platforms. | ||
| + | |||
| + | The driving biological projects for NA-MIC include the study of autism, lupus, prostate cancer, VCSF, and schizophrenia, but the methods are applicable to many other diseases. The computational tools developed in NA-MIC are used to perform image-analysis at a range of scales, and across a range of modalities including diffusion MRI, quantitative ECG, and metabolic and receptor PET, but potentially including microscopic, genomic, and other image data. It applies to image data from individual patients, and to studies executed across large populations. The data is taken from subjects across a wide range of time scales and ultimately apply to a broad range of diseases in a broad range of organs. | ||
Latest revision as of 20:35, 8 November 2008
Home < Events:August 2008 NCBC AHMContents
Agenda
There will be a all hands meeting for the NCBC program at NIH.
* The dates are August 13-14 and the location will be on the NIH campus
Abstract: The National Alliance for Medical Imaging Computing (NA-MIC) develops a collection of tools and applications to improve the deployment of advanced image analysis methods to the research community. By combining science and technology, the NA-MIC kit fosters leading edge clinical research achievements in a collaborative and open-source environment. The Science Fair will present the NA-MIC kit through the perspectives of each of our three target audiences: clinical researchers, biomedical engineers and algorithm developers. Attendees will be exposed to potential uses of the NA-MIC kit in the context of 3D visualization to facilitate data interpretation, in the use of advanced image analysis to extract relevant information, and in the development of tailored plug-ins to extend image computing capabilities. Through demonstrations of image processing methods within the 3D Slicer deployment platform, and presentations of NA-MIC pragmatic teaching initiatives to translate techniques into skills, the Science Fair will provide an overview of the benefits of NA-MIC efforts to the research community.
|
NA-MIC is one of seven National Centers for Biomedical Computing, part of the NIH roadmap initiative.
| |

| ||
NCBC Presentation Materials
Russ Altman will be giving a 10 minutes overview over the entire NCBC program. He has reqested from each of the NCBCs the following information about accomplishments.
DBPs
A short summary of the original and renewal DBPs (this will also be used in the DBP sig breakout session). Basically title and one paragraph description.
1) During the first three years of NA-MIC, Core 3 consisted of four DBPs which were grouped into two thrusts.
- Thrust 1 was directed by Drs. Shenton and Saykin. Thrust 2 was directed by Drs. Potkin and Kennedy. The focus of the research was to utilize neuroimaging tools to evaluate fronto-temporal connectivity abnormalities in schizophrenia, as well as abnormalities in hemispheric connections (i.e., corpus callosum), and abnormalities in the anterior limb of the internal capsule. Improved segmentation techniques, coregistration of structural MRI, DTI-MR, and fMRI, as well as novel processing tools for evaluating white matter fiber tracts and interregional functional connectivity were needed to accomplish these goals, and they were developed in conjunction with Cores 1 and 2.
- Findings from this project, which involve both structural and functional information about brain abnormalities in schizophrenia, were correlated with neurocognitive, clinical, and behavioral data in order to understand further the relationship between brain abnormalities and cognition/behavior in schizophrenia.
2) Starting with the 4th year of NA-MIC, the DBPs were shifted from schizophrenia to lupus, autism, velocardiofacial syndrome (VCSF), and prostate cancer.
- These DBPs now drive the computational research within NA-MIC. Specifically:
- Drs. Jeremy Bockholt and Charles Gasparovi at the MIND Institute and the University of New Mexico are analyzing brain lesions in Neuropsychiatric Systemic Lupus Erythematosis,
- Drs. Heather Hazlett and Joseph Piven at University of North Carolina, Chapel Hill are conducting a longitudinal MRI study of early brain development in Autism,
- Dr. Marek Kubicki at Harvard Medical School is investigating VCSF as a genetic model for schizophrenia, and
- Dr. Gabor Fichtinger at Queens University is developing a robotic percutaneous surgery system for treatment of prostate cancer.
Publications
1) A few key journal publications that have emerged directly from each center (say the top 5 per center). These should not be presented in a center-specific context but just to show the volume and quality of scientific production. You should be able to get these directly from your progress reports.
- We use our publications database to gather download statistics about our publications. See here for an up-to-date view.
- This are the top five papers from our database in terms of number of downloads between September 2007 and July 2008
- Pohl K, Fisher J, Bouix S, Shenton M, McCarley R, Grimson E, Kikinis R, Wells W. Using the logarithm of odds to define a vector space on probabilistic atlases. Med Image Anal. 2007 Oct;11(5):465-77.
- Hata N, Piper S, Jolesz F, Tempany C, Black P, Morikawa S, Iseki H, Hashizume M, Kikinis R. Application of Open Source Image Guided Therapy Software in MR-guided Therapies. Int Conf Med Image Comput Comput Assist Interv. 2007;10(Pt 1):491-8
- Yeo B, Sabuncu M, Desikan R, Fischl B, Golland P. Effects of Registration Regularization and Atlas Sharpness on Segmentation Accuracy. Int Conf Med Image Comput Comput Assist Interv. 2007;10(Pt 1):683-91
- Bouix S, Martin-Fernandez M, Ungar L, Nakamura M, Koo M, McCarley R, Shenton M. On evaluating brain tissue classifiers without a ground truth. Neuroimage. 2007 Jul 15;36(4):1207-1224
- Georgiou T, Michailovich O, Rathi Y, Malcolm J, Tannenbaum A. Distribution Metrics and Image Segmentation. Linear Algebra and its Applications. 2007;425(2-3):663-672
2) Some statistics of publications: total number in the last year or cumulative. Again, these should be presented in aggregate rather than by center.
- 198 cumulative publications. http://www.na-mic.org/pages/Special:Publications?collection=12
- See also here for a geographical distribution of the downloads. This link is sometimes slow.
Tools
A list of tools produced by the center with one or two highlights where you have high volume download or many journal citations.
- NA-MIC Kit, which includes 3D Slicer, a popular visualization package (see here for un-curated download statistics--it is helpful if you sort by number of downloads to see the full impact).
Outreach
- List of conferences/meetings where people in your center have explicitly advertised or demoed the NCBC’s activities.
- MICCAI, RSNA, Human Brain Mapping, IGT Workshops at NIH
- Newsletters, conferences, and other activities promoted by the centers.
- The semi-annual hands-on event called the "NA-MIC Project Week".
Collaborations
http://wiki.na-mic.org/Wiki/index.php/NA-MIC_External_Collaborations
Buiding Bridges Postdoc
- Who was the bridge built to?
- The National Center for Microscopy and Imaging Research (NCMIR; http://ncmir.ucsd.edu) at the University of California, San Diego. A key emphasis of NCMIR is the application of advanced imaging technologies to the nervous system in health and disease. Affiliated with UCSD’s Center for Research in Biological Systems (CRBS), the NCMIR is a recognized authority in the development of technologies for high throughput multi-scale imaging and analysis of biological systems at the mesoscale, the dimensional range spanning 5 nm3 and 50µm3. Macromolecules, organelles, and multi-component structures like synapses which are encompassed in this dimensional range have traditionally been challenging to study because they fall in the resolution gap between X-ray crystallography at one end and medical imaging at the other.
- What is the general topic of the research?
- Work within the Slicer environment to address multi-scale microscopy imaging data, thereby allowing for the analysis and visualization of cellular data. The project has focused on the importation of data into Slicer and the investigation of algorithms for cellular and sub-cellular data (e.g. see - http://www.na-mic.org/Wiki/index.php/Projects/Slicer3/2007_Project_Week_Support_for_electron_microscopy).
NA-MIC Description
NA-MIC is a multi-institutional, interdisciplinary team of computer scientists, software engineers, and medical investigators who develop computational tools for the analysis and visualization of medical image data. The purpose of the center is to provide the infrastructure and environment for the development of computational algorithms and open source technologies, and then oversee the training and dissemination of these tools to the medical research community. This world-class software and development environment serves as a foundation for accelerating the development and deployment of computational tools that are readily accessible to the medical research community. The team combines cutting-edge computer vision research (to create medical imaging analysis algorithms) with state of the art software engineering techniques (based on "extreme" programming techniques in a distributed, open-source environment) to enable computational examination of both basic neuroscience and neurological disorders. In developing this infrastructure resource, the team is significantly expanding upon proven open systems technology and platforms.
The driving biological projects for NA-MIC include the study of autism, lupus, prostate cancer, VCSF, and schizophrenia, but the methods are applicable to many other diseases. The computational tools developed in NA-MIC are used to perform image-analysis at a range of scales, and across a range of modalities including diffusion MRI, quantitative ECG, and metabolic and receptor PET, but potentially including microscopic, genomic, and other image data. It applies to image data from individual patients, and to studies executed across large populations. The data is taken from subjects across a wide range of time scales and ultimately apply to a broad range of diseases in a broad range of organs.