Difference between revisions of "AHM2009:Mind"
Hjbockholt (talk | contribs) |
Hjbockholt (talk | contribs) |
||
| (48 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| − | == | + | ==MRN Roadmap Project== |
{| | {| | ||
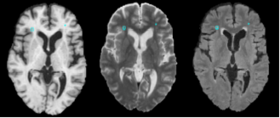
| − | |[[Image: | + | |[[Image:Lupus_acquisition.png|thumb|280px|Image Acquisition]] |
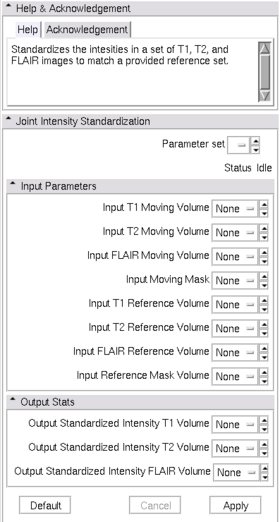
| − | |[[Image: | + | |[[Image:Intensity_standardize_view.png|thumb|280px|Slicer3 Module Image Intensity Standardization]] |
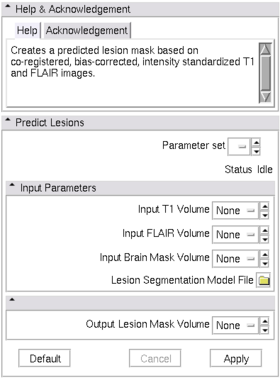
| − | |[[Image: | + | |[[Image:Predict_lesion_view.png|thumb|280px|Slicer3 Module Lesion Classification]] |
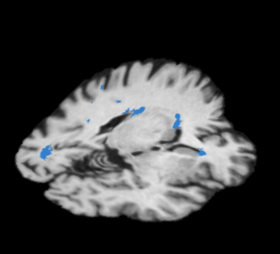
| + | |[[Image:Lupus_lesion_label_map.png|thumb|280px|Lesion Classification Result]] | ||
|} | |} | ||
== Overview == | == Overview == | ||
* What problem does the pipeline solve, and who is the targeted user? | * What problem does the pipeline solve, and who is the targeted user? | ||
| − | **The pipeline attempts to solve the problem of segmenting white matter | + | **The pipeline attempts to solve the problem of segmenting white matter lesions in Neuropsychiatric Systemic Lupus Erythematosus(NPSLE). The automated capability is aimed at clinical researchers using Slicer3 software. The utility of having accurate lesion labels permits summary of perfusion within lesions, correlation of lesion load by location with neuropsychiatric symptoms, and summary of structural image intensities or DTI scalars within lesion boundaries. |
| − | * How does the pipeline compare to state of the art | + | * How does the pipeline compare to state of the art? |
| − | **The state of the art is a work in progress in | + | **The state of the art in brain lesion classification is a work in progress in NPSLE, as well as, other disorders. There are many approaches that attempt to solve lesion classification in Multiple Sclerosis (where it is much more common to perform neuroimaging studies). Some of these approaches are automated or semi-automated; however, all automatic approaches suffer a lack of ground truth. It is difficult for human manual raters to agree on fuzzy boundaries across different image contrasts (e.g., T1, T2, FLAIR). |
| − | A good example of the | + | A good example of the challenge that remains in terms of lesion segmentation was well represented at the 2008 MICCAI Conference [http://www.ia.unc.edu/MSseg/ MS Lesion Segmentation Challenge(organized by Warfield and Styner)] this past September in the form of a grand challenge in segmentation. Several groups, including this DBP, competed in lesion segmentation contest, participants were first provided training data-sets with two manual labellings, their respective approaches were judged in an on-site, real-time competition on additional novel data sets. |
| + | |||
| + | Through participation in the recent lesion challenge, a thorough literature review of recent advances, and trying out several approaches (EM-Segment(S. Wells), k-means/bayesian(V. Magnotta), outlier detection(M. Prastawa), etc.), we think we have honed in on a novel methodology that will perform well for providing lesion maps in NPSLE; in addition, generalize well (following training) for other neurological disorders, such as MS, vascular dementia, and other disorders of the brain. | ||
==Detailed Information about the Pipeline== | ==Detailed Information about the Pipeline== | ||
| − | * Illustrate the components and workflow of the pipeline using your own data | + | *Illustrate the components and workflow of the pipeline using your own data |
| − | + | **Method | |
| − | Image: | + | ***A synergistic combination of supervised machine learning methods |
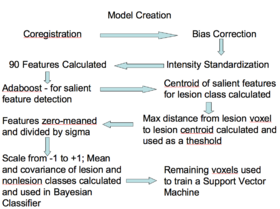
| − | + | **Training [[Image:Sle_wmls_model_generation.png|thumb|280px|Training]] | |
| − | * | + | ***Approximately 90 features were computed based on the T1, T2, and FLAIR, including neighborhood means with varying radii, mathematical morphometry dilation and erosion, kmeans clustering, and gradients, among others |
| + | ***Adaboost was applied to the data to find the 20 features that best discriminate lesion from non-lesion | ||
| + | ***Those 20 features were then calculated for all lesions, for all subjects, then zero-meaned and the standard deviation was set to one. | ||
| + | ***The centroid of the lesions was then calculated and the max distance found between the centroid and the lesion voxels | ||
| + | ***The max distance threshold was used to exclude voxels that had no chance of being lesions | ||
| + | ***The features for all voxels within the distance threshold were calculated and scaled to a range of negative one to positive one | ||
| + | ***The means and covariance of these features were calculated for both the lesion and non-lesion classes and used to define the two classes in a Bayesian classifier | ||
| + | ***A parameter search was then performed to find the prior that gave the best combination of Specificity and Sensitivity. | ||
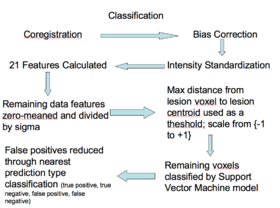
| + | **Classifying [[Image:Sle_wmls_model_creation.png|thumb|280px|Classifying]] | ||
| + | ***The 20 relevant features are calculated, zero-meaned and sigma set to one, thresholded based on the distance to the lesion centroid, and then passed to the Bayesian classifier | ||
==Software & documentation== | ==Software & documentation== | ||
| − | * [http://www.nitrc.org/frs/download.php/570/LesionSegmentationApplications.tgz tutorial software] | + | *We have a very active project for this pipeline on the NITRC resource [http://www.nitrc.org/frs/?group_id=180 3DSlicerLupusLesionModule][[Image:Lupus_nitrc_weekly_stat.png|thumb|175px|Image Acquisition]] |
| − | * [http://www.nitrc.org/frs/download.php/569/LesionSegmentationTutorialData.tgz tutorial data] | + | **[http://www.nitrc.org/frs/download.php/570/LesionSegmentationApplications.tgz tutorial software] |
| − | * [http://www.nitrc.org/frs/download.php/571/Slicer3Training_WhiteMatterLesions_v2.0.ppt.pdf | + | **[http://www.nitrc.org/frs/download.php/569/LesionSegmentationTutorialData.tgz tutorial data] |
| + | **[http://www.nitrc.org/frs/download.php/571/Slicer3Training_WhiteMatterLesions_v2.0.ppt.pdf end-user tutorial] | ||
==Team== | ==Team== | ||
| − | * DBP: | + | * DBP: H Jeremy Bockholt (PI), Charles Gasparovic(co-PI), Mark Scully(Engineer), The Mind Research Network |
| − | * Core 1: | + | * Core 1: Ross Whitaker, University of Utah |
| − | * Core 2: | + | * Core 2: Steve Pieper, Isomics |
| + | * Consultant: Vincent Magnotta, University of Iowa | ||
* Contact: H. Jeremy Bockholt, hjbockholt@mrn.org | * Contact: H. Jeremy Bockholt, hjbockholt@mrn.org | ||
| Line 43: | Line 57: | ||
* [http://wiki.na-mic.org/Wiki/index.php/NA-MIC_Lupus_DBP_Worskhop_SFN2008 2008 SFN conference outreach activity] | * [http://wiki.na-mic.org/Wiki/index.php/NA-MIC_Lupus_DBP_Worskhop_SFN2008 2008 SFN conference outreach activity] | ||
| − | * 2009 AAN conference outreach activity | + | ** 11 interchanges with clinical neuroscientist researchers |
| − | + | ** 2 users have followed up with questions on poster and software tool/data | |
| + | * April 25 - May 2 2009 AAN conference outreach activity | ||
==References== | ==References== | ||
| − | Adaboost and Support Vector Machines for White Matter Lesion | + | #Adaboost and Support Vector Machines for White Matter Lesion Segmentation in MR Images. Quddus A, Fieguth P, Basir O. PAMI Lab, Department of Electrical and Computer Engineering, University of Waterloo, Waterloo, ON, Canada. Conf Proc IEEE Eng Med Biol Soc. 2005;1:463-6. http://www.ncbi.nlm.nih.gov/pubmed/17282216 |
| − | Segmentation in MR Images. | + | #Architecture for an Artificial Immune System Steven A. Hofmeyr, Stephanie Forrest Evolutionary Computation Winter 2000, Vol. 8, No. 4, Pages 443-473 http://www.mitpressjournals.org/doi/abs/10.1162/106365600568257 |
| − | Quddus A, Fieguth P, Basir O. | + | #Automated segmentation of white matter lesions in 3D brain MR images, using multivariate pattern classification Zhiqiang Lao; Dinggang Shen; Jawad, A.; Karacali, B.; Dengfeng Liu; Melhem, E.R.; Bryan, R.N.; Davatzikos, C. Biomedical Imaging: Nano to Macro, 2006. 3rd IEEE International Symposium on Volume , Issue , 6-9 April 2006 Page(s): 307 - 310 http://ieeexplore.ieee.org/Xplore/login.jsp?url=/iel5/10818/34114/01624914.pdf?temp=x |
| − | PAMI Lab, Department of Electrical and Computer Engineering, | + | #Automatic Segmentation of MS Lesions Using a Contextual Model for the MICCAI Grand Challenge Jonathan H. Morra, Zhuowen Tu, Arthur W. Toga, Paul M. Thompson IJ - 2008 MICCAI Workshop - MS Lesion Segmentation http://www.midasjournal.org/browse/publication/280 |
| − | University of Waterloo, Waterloo, ON, Canada. | ||
| − | Conf Proc IEEE Eng Med Biol Soc. 2005;1:463-6. | ||
| − | http://www.ncbi.nlm.nih.gov/pubmed/17282216 | ||
| − | |||
| − | Architecture for an Artificial Immune System | ||
| − | Steven A. Hofmeyr, Stephanie Forrest | ||
| − | Evolutionary Computation Winter 2000, Vol. 8, No. 4, Pages 443-473 | ||
| − | http://www.mitpressjournals.org/doi/abs/10.1162/106365600568257 | ||
| − | |||
| − | Automated segmentation of white matter lesions in 3D brain MR images, | ||
| − | using multivariate pattern classification | ||
| − | Zhiqiang Lao; Dinggang Shen; Jawad, A.; Karacali, B.; Dengfeng Liu; | ||
| − | Melhem, E.R.; Bryan, R.N.; Davatzikos, C. | ||
| − | Biomedical Imaging: Nano to Macro, 2006. 3rd IEEE International Symposium on | ||
| − | Volume , Issue , 6-9 April 2006 Page(s): 307 - 310 | ||
| − | http://ieeexplore.ieee.org/Xplore/login.jsp?url=/iel5/10818/34114/01624914.pdf?temp=x | ||
| − | |||
| − | Automatic Segmentation of MS Lesions Using a Contextual Model for the | ||
| − | MICCAI Grand Challenge | ||
| − | Jonathan H. Morra, Zhuowen Tu, Arthur W. Toga, Paul M. Thompson | ||
| − | IJ - 2008 MICCAI Workshop - MS Lesion Segmentation | ||
| − | http://www.midasjournal.org/browse/publication/280 | ||
Latest revision as of 15:35, 8 January 2009
Home < AHM2009:Mind
MRN Roadmap Project
Overview
- What problem does the pipeline solve, and who is the targeted user?
- The pipeline attempts to solve the problem of segmenting white matter lesions in Neuropsychiatric Systemic Lupus Erythematosus(NPSLE). The automated capability is aimed at clinical researchers using Slicer3 software. The utility of having accurate lesion labels permits summary of perfusion within lesions, correlation of lesion load by location with neuropsychiatric symptoms, and summary of structural image intensities or DTI scalars within lesion boundaries.
- How does the pipeline compare to state of the art?
- The state of the art in brain lesion classification is a work in progress in NPSLE, as well as, other disorders. There are many approaches that attempt to solve lesion classification in Multiple Sclerosis (where it is much more common to perform neuroimaging studies). Some of these approaches are automated or semi-automated; however, all automatic approaches suffer a lack of ground truth. It is difficult for human manual raters to agree on fuzzy boundaries across different image contrasts (e.g., T1, T2, FLAIR).
A good example of the challenge that remains in terms of lesion segmentation was well represented at the 2008 MICCAI Conference MS Lesion Segmentation Challenge(organized by Warfield and Styner) this past September in the form of a grand challenge in segmentation. Several groups, including this DBP, competed in lesion segmentation contest, participants were first provided training data-sets with two manual labellings, their respective approaches were judged in an on-site, real-time competition on additional novel data sets.
Through participation in the recent lesion challenge, a thorough literature review of recent advances, and trying out several approaches (EM-Segment(S. Wells), k-means/bayesian(V. Magnotta), outlier detection(M. Prastawa), etc.), we think we have honed in on a novel methodology that will perform well for providing lesion maps in NPSLE; in addition, generalize well (following training) for other neurological disorders, such as MS, vascular dementia, and other disorders of the brain.
Detailed Information about the Pipeline
- Illustrate the components and workflow of the pipeline using your own data
- Method
- A synergistic combination of supervised machine learning methods
- Training
- Approximately 90 features were computed based on the T1, T2, and FLAIR, including neighborhood means with varying radii, mathematical morphometry dilation and erosion, kmeans clustering, and gradients, among others
- Adaboost was applied to the data to find the 20 features that best discriminate lesion from non-lesion
- Those 20 features were then calculated for all lesions, for all subjects, then zero-meaned and the standard deviation was set to one.
- The centroid of the lesions was then calculated and the max distance found between the centroid and the lesion voxels
- The max distance threshold was used to exclude voxels that had no chance of being lesions
- The features for all voxels within the distance threshold were calculated and scaled to a range of negative one to positive one
- The means and covariance of these features were calculated for both the lesion and non-lesion classes and used to define the two classes in a Bayesian classifier
- A parameter search was then performed to find the prior that gave the best combination of Specificity and Sensitivity.
- Classifying
- The 20 relevant features are calculated, zero-meaned and sigma set to one, thresholded based on the distance to the lesion centroid, and then passed to the Bayesian classifier
- Method
Software & documentation
- We have a very active project for this pipeline on the NITRC resource 3DSlicerLupusLesionModule
Team
- DBP: H Jeremy Bockholt (PI), Charles Gasparovic(co-PI), Mark Scully(Engineer), The Mind Research Network
- Core 1: Ross Whitaker, University of Utah
- Core 2: Steve Pieper, Isomics
- Consultant: Vincent Magnotta, University of Iowa
- Contact: H. Jeremy Bockholt, hjbockholt@mrn.org
Outreach
- Publication Links to the PubDB.
- H. J. Bockholt, V. A. Magnotta, M. Scully, C. Gasparovic, B. Davis, K. Pohl, R. Whitaker, S. Pieper, C. Roldan, R. Jung, R. Hayek, W. Sibbitt, J. Sharrar, P. Pellegrino, R. Kikinis. A novel automated method for classification of white matter lesions in systemic lupus erythematosus. Presented at the 38th annual meeting of the Society for Neuroscience, Washingto, DC, 15 – 19 November 2008
- Scully M., Magnotta V., Gasparovic C., Pelligrimo P., Feis D., Bockholt H.J. 3D Segmentation In The Clinic: A Grand Challenge II at MICCAI 2008 - MS Lesion Segmentation. IJ - 2008 MICCAI Workshop - MS Lesion Segmentation. Available http://grand-challenge2008.bigr.nl/proceedings/pdfs/msls08/282_Scully.pdf
- 2008 SFN conference outreach activity
- 11 interchanges with clinical neuroscientist researchers
- 2 users have followed up with questions on poster and software tool/data
- April 25 - May 2 2009 AAN conference outreach activity
References
- Adaboost and Support Vector Machines for White Matter Lesion Segmentation in MR Images. Quddus A, Fieguth P, Basir O. PAMI Lab, Department of Electrical and Computer Engineering, University of Waterloo, Waterloo, ON, Canada. Conf Proc IEEE Eng Med Biol Soc. 2005;1:463-6. http://www.ncbi.nlm.nih.gov/pubmed/17282216
- Architecture for an Artificial Immune System Steven A. Hofmeyr, Stephanie Forrest Evolutionary Computation Winter 2000, Vol. 8, No. 4, Pages 443-473 http://www.mitpressjournals.org/doi/abs/10.1162/106365600568257
- Automated segmentation of white matter lesions in 3D brain MR images, using multivariate pattern classification Zhiqiang Lao; Dinggang Shen; Jawad, A.; Karacali, B.; Dengfeng Liu; Melhem, E.R.; Bryan, R.N.; Davatzikos, C. Biomedical Imaging: Nano to Macro, 2006. 3rd IEEE International Symposium on Volume , Issue , 6-9 April 2006 Page(s): 307 - 310 http://ieeexplore.ieee.org/Xplore/login.jsp?url=/iel5/10818/34114/01624914.pdf?temp=x
- Automatic Segmentation of MS Lesions Using a Contextual Model for the MICCAI Grand Challenge Jonathan H. Morra, Zhuowen Tu, Arthur W. Toga, Paul M. Thompson IJ - 2008 MICCAI Workshop - MS Lesion Segmentation http://www.midasjournal.org/browse/publication/280