Difference between revisions of "2015 Summer Project Week:LungCAD"
From NAMIC Wiki
| Line 33: | Line 33: | ||
* Completed analysis for 248 GGOs and trained SVM with an accuracy of 89% to classify GGOs | * Completed analysis for 248 GGOs and trained SVM with an accuracy of 89% to classify GGOs | ||
* Developed the framework for the LungCAD module in Slicer | * Developed the framework for the LungCAD module in Slicer | ||
| − | * | + | * Evaluated the Lesion segmentation algorithm as part of the Chest Imaging Platform |
* Segmentation works well and is able to prevent the segmentation of vessels running through the lesion | * Segmentation works well and is able to prevent the segmentation of vessels running through the lesion | ||
* LungCAD will call Lesion Segmentation CLI for segmentation and HeterogeneityCAD to evaluate features | * LungCAD will call Lesion Segmentation CLI for segmentation and HeterogeneityCAD to evaluate features | ||
Revision as of 14:41, 24 June 2015
Home < 2015 Summer Project Week:LungCADKey Investigators
- Jayender Jagadeesan
- Tobias Penskofer
- Sandy Wells
- Clara Meiner
- Raul San Jose Estepar
Project Description
Objective
- Develop a module in 3D Slicer to segment the ground glass opacity (GGO) tumor, apply HeterogeneityCAD to obtain imaging metrics and classify the GGO.
- Provide the module as an extension part of OpenCAD
Approach, Plan
- Implement a simple region growing algorithm
- Apply HeterogeneityCAD module
- Use predetermined SVM classifier to decide the lesion type
Progress
- Completed analysis for 248 GGOs and trained SVM with an accuracy of 89% to classify GGOs
- Developed the framework for the LungCAD module in Slicer
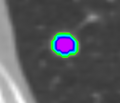
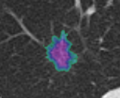
- Evaluated the Lesion segmentation algorithm as part of the Chest Imaging Platform
- Segmentation works well and is able to prevent the segmentation of vessels running through the lesion
- LungCAD will call Lesion Segmentation CLI for segmentation and HeterogeneityCAD to evaluate features
- Pre-processed SVM will be utilized to classify the GGOs