Difference between revisions of "2009 Summer Project Week 4D Imaging"
From NAMIC Wiki
| (18 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Project Week Main Page]] |
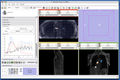
| − | Image: | + | Image:3DSlicerFourDAnalysis_Screenshot.png|Screen shot of 4D Analysis Module |
| − | |||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
* BWH: Junichi Tokuda, Wendy Plesniak, Nobuhiko Hata | * BWH: Junichi Tokuda, Wendy Plesniak, Nobuhiko Hata | ||
| − | * | + | * WFU:Craig A. Hamilton |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 14: | Line 13: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Implement a set of 3D Slicer modules to handle 4D images in 3D Slicer for perfusion analysis, cardiac, etc. | |
| + | including: | ||
| + | *Handling | ||
| + | **Loading 4D volume | ||
| + | **Scroll time-line | ||
| + | **4D image editing | ||
| + | **Recording [[2009_Summer_Project_Week_4D_Gated_US_In_Slicer| 4D Gated US]] | ||
| + | *Processing | ||
| + | **Image registration for motion compensation | ||
| + | *Analysis | ||
| + | **Perfusion analysis: fitting pharmacokinetic model | ||
| + | |||
| + | *We are going to organize 4D imaging session during the meeting. | ||
| + | Please join us, if you are interested in. | ||
| Line 22: | Line 34: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | We will work on the following tasks: | |
| − | + | *Implementation of '''4D Image''' module that provides: | |
| + | **4D image loading: Loading a series of 3D images from a specified director. The data can be either in DICOM or NRRD format. | ||
| + | **Time line scroll-bar interface: scrolling the frame in time-direction. It allows you to scroll the frame for foreground and background screens independently to compare two images at the different time points. | ||
| + | **Frame editing: Reorganizing the time series data (optional) | ||
| + | *Implementation of '''4D Analysis''' module that provides: | ||
| + | **Intensity plot: Plotting temporal changes of intensities at specified regions. This feature is useful for analyzing dynamic contrast images. | ||
| + | **Model fitting: A python interface to analyze intensity curves obtained from the 4D images. The interface is useful to fit pharmacokinetic models to intensity curves to obtain perfusion parameters. | ||
| + | *Investigating BatchMake as an infrastructure for time-series image processing. | ||
| + | **4D Cropping: Cropping volumes in a time-series data using BatchMake. | ||
| + | **4D Image registration: Registering each volume frame to a key-frame to compensate organ motion. | ||
| + | **Image registration using cluster (Optional) | ||
</div> | </div> | ||
| Line 30: | Line 52: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | Before the project week, we have completed: | |
| + | *Initial implementation of 4D Image module. The module is available at http://svn.slicer.org/Slicer3/trunk/Modules/FourDImage | ||
| + | *Initial implementation of 4D Analysis module. The Python interface has to be fixed. The module is available at http://svn.slicer.org/Slicer3/trunk/Modules/FourDAnalysis | ||
| + | *[[2009_Summer_Project_Week_4D_Imaging_discussion| 4D Meeting]] was organized. | ||
| + | **Demo of 4D module | ||
| + | **4D Bundle | ||
| + | **Application | ||
| + | **Memory usage | ||
| + | **Pipeline processing | ||
| + | |||
</div> | </div> | ||
| Line 36: | Line 67: | ||
==References== | ==References== | ||
| − | + | ||
| − | + | The preliminary module implementation is described in [[Slicer3:FourDAnalysis]]. | |
| − | |||
| − | |||
Latest revision as of 14:40, 26 June 2009
Home < 2009 Summer Project Week 4D ImagingKey Investigators
- BWH: Junichi Tokuda, Wendy Plesniak, Nobuhiko Hata
- WFU:Craig A. Hamilton
Objective
Implement a set of 3D Slicer modules to handle 4D images in 3D Slicer for perfusion analysis, cardiac, etc. including:
- Handling
- Loading 4D volume
- Scroll time-line
- 4D image editing
- Recording 4D Gated US
- Processing
- Image registration for motion compensation
- Analysis
- Perfusion analysis: fitting pharmacokinetic model
- We are going to organize 4D imaging session during the meeting.
Please join us, if you are interested in.
Approach, Plan
We will work on the following tasks:
- Implementation of 4D Image module that provides:
- 4D image loading: Loading a series of 3D images from a specified director. The data can be either in DICOM or NRRD format.
- Time line scroll-bar interface: scrolling the frame in time-direction. It allows you to scroll the frame for foreground and background screens independently to compare two images at the different time points.
- Frame editing: Reorganizing the time series data (optional)
- Implementation of 4D Analysis module that provides:
- Intensity plot: Plotting temporal changes of intensities at specified regions. This feature is useful for analyzing dynamic contrast images.
- Model fitting: A python interface to analyze intensity curves obtained from the 4D images. The interface is useful to fit pharmacokinetic models to intensity curves to obtain perfusion parameters.
- Investigating BatchMake as an infrastructure for time-series image processing.
- 4D Cropping: Cropping volumes in a time-series data using BatchMake.
- 4D Image registration: Registering each volume frame to a key-frame to compensate organ motion.
- Image registration using cluster (Optional)
Progress
Before the project week, we have completed:
- Initial implementation of 4D Image module. The module is available at http://svn.slicer.org/Slicer3/trunk/Modules/FourDImage
- Initial implementation of 4D Analysis module. The Python interface has to be fixed. The module is available at http://svn.slicer.org/Slicer3/trunk/Modules/FourDAnalysis
- 4D Meeting was organized.
- Demo of 4D module
- 4D Bundle
- Application
- Memory usage
- Pipeline processing
References
The preliminary module implementation is described in Slicer3:FourDAnalysis.