Difference between revisions of "2009 Summer Project Week Liver Ablation Slicer"
From NAMIC Wiki
| (12 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week| | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Projects List]] |
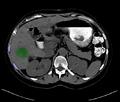
| + | Image:originalSegmentation.png|Segmentation showing no pass zones (ribs)and tumor. | ||
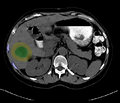
| + | Image:segmentationAfterDilation.png|Segmented tumor region is dilated according to physician prescribed ablation margin. | ||
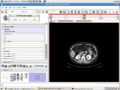
| + | Image:Slicer_liver_module.png|IGT planning module for liver albation in Slicer3. | ||
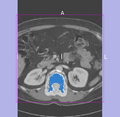
| + | Image:Segmentation_dilated_by_slicer.png|Segmented tumor region is dilated in Slicer3. | ||
</gallery> | </gallery> | ||
| Line 12: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Implement a complete workflow: | |
| + | #Load data. | ||
| + | #Manually mark regions (tumor, entry, critical structure). | ||
| + | #Process segmentation and export information to planning module (executable plugin with command line options). | ||
| + | #Load results of the optimization program. | ||
| + | #Configure OpenIGTLink module on Slicer and run OpenIGTLink IGSTK client. | ||
| + | #Navigate. | ||
</div> | </div> | ||
| Line 18: | Line 28: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | #Integrate code from Georgetown for step 3 into Slicer. | |
| − | + | #Decide on appropriate format for describing the output of the optimization (set of trajectories and ablations along each trajectory). | |
| − | + | #Implement a stub executable plugin as a stand in for the optimization program. | |
| + | #Test the integrated workflow. | ||
</div> | </div> | ||
| Line 26: | Line 37: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Completed GUI design and implementation for "planning" part of the workflow. | |
| + | * An unique feature in the planning part were tumor segmentation and ablation volume planning, taking ablation magin into account. | ||
| + | * "Navigation" part 80% done. | ||
| + | * In "Navigation", "IGT Guidance" widget was designed and implemented to be shared with other IGT investigators. | ||
| + | |||
</div> | </div> | ||
| + | </div> | ||
| + | |||
| + | <div style="width: 97%; float: left;"> | ||
| + | |||
| + | ==References== | ||
| + | *Z. Yaniv, E. Wilson, D. Lindisch, K. Cleary, "Electromagnetic Tracking in the Clinical Environment", Med. Phys., Vol. 36(3), pp. 876-892, 2009. PMID: 19378748 | ||
| + | *J. Tokuda et al., "OpenIGTLink: An open network protocol for image- guided therapy environment," International Journal of Medical Robotics and Computer Assisted Surgery, to appear. | ||
| + | |||
</div> | </div> | ||
Latest revision as of 15:16, 26 June 2009
Home < 2009 Summer Project Week Liver Ablation SlicerKey Investigators
- BWH: Haiying Liu, Noby Hata
- Georgetown: Ziv Yaniv
Objective
Implement a complete workflow:
- Load data.
- Manually mark regions (tumor, entry, critical structure).
- Process segmentation and export information to planning module (executable plugin with command line options).
- Load results of the optimization program.
- Configure OpenIGTLink module on Slicer and run OpenIGTLink IGSTK client.
- Navigate.
Approach, Plan
- Integrate code from Georgetown for step 3 into Slicer.
- Decide on appropriate format for describing the output of the optimization (set of trajectories and ablations along each trajectory).
- Implement a stub executable plugin as a stand in for the optimization program.
- Test the integrated workflow.
Progress
- Completed GUI design and implementation for "planning" part of the workflow.
- An unique feature in the planning part were tumor segmentation and ablation volume planning, taking ablation magin into account.
- "Navigation" part 80% done.
- In "Navigation", "IGT Guidance" widget was designed and implemented to be shared with other IGT investigators.
References
- Z. Yaniv, E. Wilson, D. Lindisch, K. Cleary, "Electromagnetic Tracking in the Clinical Environment", Med. Phys., Vol. 36(3), pp. 876-892, 2009. PMID: 19378748
- J. Tokuda et al., "OpenIGTLink: An open network protocol for image- guided therapy environment," International Journal of Medical Robotics and Computer Assisted Surgery, to appear.