Difference between revisions of "2009 Summer Project Week Liver Ablation Slicer"
From NAMIC Wiki
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week| | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Projects List]] |
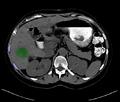
Image:originalSegmentation.png|Segmentation showing no pass zones (ribs)and tumor. | Image:originalSegmentation.png|Segmentation showing no pass zones (ribs)and tumor. | ||
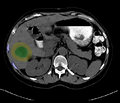
Image:segmentationAfterDilation.png|Segmented tumor region is dilated according to physician prescribed ablation margin. | Image:segmentationAfterDilation.png|Segmented tumor region is dilated according to physician prescribed ablation margin. | ||
Revision as of 02:07, 17 June 2009
Home < 2009 Summer Project Week Liver Ablation SlicerKey Investigators
- BWH: Haiying Liu, Noby Hata
- Georgetown: Ziv Yaniv
Objective
Implement a complete workflow:
- Load data.
- Manually mark regions (tumor, entry, critical structure).
- Process segmentation and export information to planning module (executable plugin with command line options).
- Load results of the optimization program.
- Configure OpenIGTLink module on Slicer and run OpenIGTLink IGSTK client.
- Navigate.
Approach, Plan
- Integrate code from Georgetown for step 3 into Slicer.
- Decide on appropriate format for describing the output of the optimization (set of trajectories and ablations along each trajectory).
- Implement a stub executable plugin as a stand in for the optimization program.
- Test the integrated workflow.
Progress
A new module is being created in Slicer3.
References
- Z. Yaniv, E. Wilson, D. Lindisch, K. Cleary, "Electromagnetic Tracking in the Clinical Environment", Med. Phys., Vol. 36(3), pp. 876-892, 2009. PMID: 19378748
- J. Tokuda et al., "OpenIGTLink: An open network protocol for image- guided therapy environment," International Journal of Medical Robotics and Computer Assisted Surgery, to appear.