Difference between revisions of "2009 Summer Project Week Lupus Lesion Segmentation"
From NAMIC Wiki
(Changed wording of wizard progress) |
|||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week| | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Projects List]] |
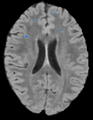
| − | Image:lesionManual-PW2009.png| | + | Image:lesionManual-PW2009.png|Manual tracing of lupus lesions. |
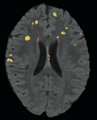
| + | Image:lesionHeatMap-PW2009.png|Predicted Heat Map. | ||
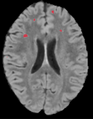
| + | Image:lesionThreshold-PW2009.png|Heat Map after thresholding. | ||
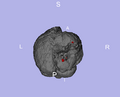
| + | Image:LesionInBrain-PW2009.png|Lesion model inside brain. | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * H. Jeremy Bockholt | + | * MRN: H. Jeremy Bockholt, Mark Scully |
| − | + | * Isomics: Steve Pieper | |
| − | * Steve Pieper | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 14: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| + | To create an end-to-end Lupus lesion segmentation and analysis pipeline that can work in both an automated and semi-supervised fashion. | ||
| + | See our [[DBP2:MIND:Roadmap| Roadmap]] for more details. | ||
</div> | </div> | ||
| Line 21: | Line 25: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| + | Our approach involves the combination of morphometric features with a non-linear classifier in order to capture more of the gestalt properties a human rater uses when segmenting Lupus lesions. | ||
| + | Our plan for the project week is to: | ||
| + | * Work with Steve Pieper to implement the wizard interface to the lesion analysis module to make the segmentation process easier. | ||
| + | * Refactor the lesion analysis code and integrate it with the Slicer code repository so it is built with Slicer. | ||
| + | * Meet with the white matter lesion segmentation group to discuss overlap. | ||
</div> | </div> | ||
| Line 28: | Line 37: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Charted the logic of the wizard interface and created a development plan with Steve Pieper. | |
| − | + | * Made the lesion segmentation module available as a downloadable extension. This obviates the need to add the source code to the Slicer repository. | |
| + | * Met with the white matter lesion segmentation project group from UNC / GE to exchange data and discuss methods. | ||
| + | * Updated tutorial reflecting feedback from dissemination core. | ||
| + | * Improved testing coverage of lesion segmentation tools. | ||
</div> | </div> | ||
</div> | </div> | ||
<div style="width: 970%; float: left;"> | <div style="width: 970%; float: left;"> | ||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 13:54, 26 June 2009
Home < 2009 Summer Project Week Lupus Lesion SegmentationKey Investigators
- MRN: H. Jeremy Bockholt, Mark Scully
- Isomics: Steve Pieper
Objective
To create an end-to-end Lupus lesion segmentation and analysis pipeline that can work in both an automated and semi-supervised fashion.
See our Roadmap for more details.
Approach, Plan
Our approach involves the combination of morphometric features with a non-linear classifier in order to capture more of the gestalt properties a human rater uses when segmenting Lupus lesions.
Our plan for the project week is to:
- Work with Steve Pieper to implement the wizard interface to the lesion analysis module to make the segmentation process easier.
- Refactor the lesion analysis code and integrate it with the Slicer code repository so it is built with Slicer.
- Meet with the white matter lesion segmentation group to discuss overlap.
Progress
- Charted the logic of the wizard interface and created a development plan with Steve Pieper.
- Made the lesion segmentation module available as a downloadable extension. This obviates the need to add the source code to the Slicer repository.
- Met with the white matter lesion segmentation project group from UNC / GE to exchange data and discuss methods.
- Updated tutorial reflecting feedback from dissemination core.
- Improved testing coverage of lesion segmentation tools.