Difference between revisions of "2009 Summer Project Week Lupus Lesion Segmentation"
From NAMIC Wiki
(Changed key investigators format) |
(Added objective and approach) |
||
| Line 15: | Line 15: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| + | To create an end-to-end Lupus lesion segmentation and analysis pipeline that can work in both an automated and semi-supervised fashion. | ||
| + | See our [[DBP2:MIND:Roadmap| Roadmap]] for more details. | ||
</div> | </div> | ||
| Line 22: | Line 24: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| + | Our approach involves the combination of morphometric features with a non-linear classifier in order to capture more of the gestalt properties a human rater uses when segmenting Lupus lesions. | ||
| + | Our plan for the project week is to: | ||
| + | * Work with Steve Pieper to implement the wizard interface to the lesion analysis module to make the segmentation process easier. | ||
| + | * Integrate the output of the lesion analysis module with change tracker to allow for longitudinal analysis. | ||
| + | * Refactor the lesion analysis code and integrate it with the Slicer code repository so it is built with Slicer. | ||
</div> | </div> | ||
Revision as of 17:23, 11 June 2009
Home < 2009 Summer Project Week Lupus Lesion SegmentationKey Investigators
- MRN: H. Jeremy Bockholt, Mark Scully
- Isomics: Steve Pieper
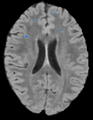
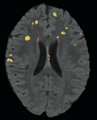
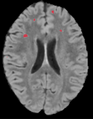
Objective
To create an end-to-end Lupus lesion segmentation and analysis pipeline that can work in both an automated and semi-supervised fashion.
See our Roadmap for more details.
Approach, Plan
Our approach involves the combination of morphometric features with a non-linear classifier in order to capture more of the gestalt properties a human rater uses when segmenting Lupus lesions.
Our plan for the project week is to:
- Work with Steve Pieper to implement the wizard interface to the lesion analysis module to make the segmentation process easier.
- Integrate the output of the lesion analysis module with change tracker to allow for longitudinal analysis.
- Refactor the lesion analysis code and integrate it with the Slicer code repository so it is built with Slicer.
Progress