Difference between revisions of "2009 Summer Project Week Slicer3 Brainlab Introduction"
From NAMIC Wiki
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | ||
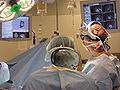
| + | Image:Slicer_dti_seeding_or.jpg|Slicer3 in OR for fiducial seeding of DTI tractography | ||
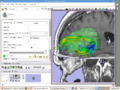
| + | Image:Slicer_dti_seeding.png|Slicer3 fiducial seeding of DTI tractography | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * BWH: Haiying Liu, Isaiah Norton |
| − | * | + | * UCLA: Nathan Hageman |
| − | |||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Demonstrate how we integrate BrainLab, BioImage Suite and Slicer3 to perform fiducial seeding of DTI tractography in Slicer3. The demo will be happening in BWH OR. | |
</div> | </div> | ||
| Line 19: | Line 20: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | Slicer3, BioImage Suite and BrainLab will be connected through a router in BWH OR. BioImage Suite will be a "bridge" between Slicer3 and BrainLab for data communication. | |
| − | |||
| − | |||
</div> | </div> | ||
| Line 27: | Line 26: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | The integrated system has been used in BWH OR for 6 neurosurgical cases. | |
</div> | </div> | ||
</div> | </div> | ||
Revision as of 19:17, 3 June 2009
Home < 2009 Summer Project Week Slicer3 Brainlab IntroductionKey Investigators
- BWH: Haiying Liu, Isaiah Norton
- UCLA: Nathan Hageman
Objective
Demonstrate how we integrate BrainLab, BioImage Suite and Slicer3 to perform fiducial seeding of DTI tractography in Slicer3. The demo will be happening in BWH OR.
Approach, Plan
Slicer3, BioImage Suite and BrainLab will be connected through a router in BWH OR. BioImage Suite will be a "bridge" between Slicer3 and BrainLab for data communication.
Progress
The integrated system has been used in BWH OR for 6 neurosurgical cases.