Difference between revisions of "2010 Summer Project Week Cortical Thickness Analysis"
From NAMIC Wiki
| Line 11: | Line 11: | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * UNC: | + | * UNC: Yundi Shi, Martin Styner |
| − | + | * Emory: Mar Sanchez | |
| − | |||
| − | |||
<h3>Objective</h3> | <h3>Objective</h3> | ||
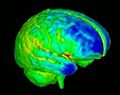
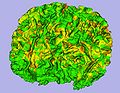
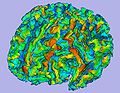
| − | + | To build an open-source software (mainly involving Slicer3) based pipeline for primate imaging, including skull stripping of T1,T2 and DTI images, tissue segmentation, cortical thickness analysis, atlas building and fiber tracing and tract-based analysis. This is part of a longitudinal study of rhesus macaque monkeys | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 34: | Line 23: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
Revision as of 20:38, 16 June 2010
Home < 2010 Summer Project Week Cortical Thickness Analysis
Key Investigators
- UNC: Yundi Shi, Martin Styner
- Emory: Mar Sanchez
Objective
To build an open-source software (mainly involving Slicer3) based pipeline for primate imaging, including skull stripping of T1,T2 and DTI images, tissue segmentation, cortical thickness analysis, atlas building and fiber tracing and tract-based analysis. This is part of a longitudinal study of rhesus macaque monkeys
Approach, Plan
Progress
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a:
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline - YES
- Extension -- loadable
- Other (Please specify)
References
- I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print.
- H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009.
- C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2010 abstract
- C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract